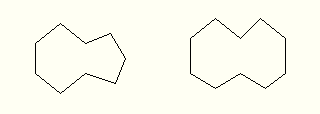Daylight's Depictions and JavaDepict
 To gain perspective let us consider the depiction problem
in general.
To gain perspective let us consider the depiction problem
in general.
-
Graph layout has been approached in 2 ways
-
The procedural approach
-
Emphasis on getting a fast algorithm even if not ideal or
theoretically well founded
-
The declarative approach
-
Layout modeled on some sort of physical force model where
aesthetics are analogs of features of that model.
-
As it involves numerical optimization of some sort, it admits
conflicting aesthetics better
-
Can be modified to allow user to determine layout rules of
his own
-
Usually leads to slower solutions
-
Daylight's system is of the second type. However it lacks
the capabilities to add chemical layout rules that a chemist or an organization
might want. i.e. it cannot be tailored
-
This problem can be dealt with at 2 levels:
-
Capture general deficiencies in depictions that chemists
would like to have addressed.
-
e.g. draw linear bonds at 120 degrees
-
Label H20
as H2O, not as OH2
-
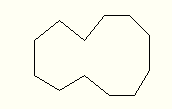
draw large cycles as combinations of 6, 7, 8 and 9 sided
polygons
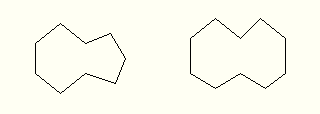
Interestingly tackling these issues is not cpu intensive,
and requires that the deficiencies be captured correctly
-
Template based preferences:
-
Allow for Templates, which will set relative bond angles
for SMARTS matches
-
Allow for specification of groups to fold and show as a single
group
-
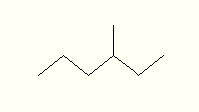
e.g. for x > 7 CH2's
in a string, fold and represent as (CH2)x
-
show -CH(CH3)2
as -Val
-
Allow for layout rules, which can be rules to set when combinations
of SMARTS appear
-
e.g. always show a polypeptide pattern N-terminal to C-terminal
with the side groups with the amino-acid names.
These issues can be cpu-intensive especially as the number
of such specifications increases