Daylight MUG 2000
February 22-25, 2000, Santa Fe, NM
ABSTRACT
We have developed a system for rapid assembly and searching of 3D-searchable
databases of ring templates taken from our corporate database and other
sources
of chemical structures. The database is generated using Daylight Toolkit
programs,
and is searched using a program called SAM, based on a published 3D
similarity
method (Atom Mapping). The system is designed to be useful for finding
novel
scaffolds for breaking out of known series, and to compare proposed
library
diversity when only a scaffold is available. We shall present an explanation
of the
method and examples of its use.
Breaking out of existing series
New combinatorial templates
Patent-busting
Use an existing scaffold as a query
Similarity Search a database of possible scaffolds using this query
We decided to use 3D databases and queries
We wanted scaffolds that would position sidechains in similar ways to a query scaffold
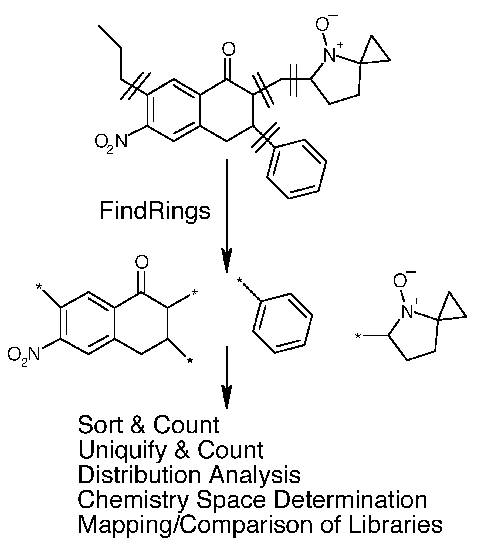
1. Chemical structures in a 2D database are processed to identify ring systems (e.g. MDDR, corporate database)2. All substituents are removed except
H atoms3. Retain substituent positions as attachment points ('*' in Smiles)
Double or triple bonded atoms
Hetero-hetero single-bonded atoms
Charged atoms attached to charged ring atoms4. Convert molecules to 3D MOL format (attachment points represented by dummy atom)
We can do this very neatly using Daylight Contrib RingSmi program (written by Jeremy Yang) for extracting ring systems, and Concord for generating 3D structures:

Define current scaffold with attachment points
Represent scaffold as SMILES with * for attachment points
Generate 3D structure with CONCORD, using Dummy atoms as attachment points
Uses a program called SAM based on the Atom Mapping method (Pepperrell, Taylor & Willett). This fits our problem well:- Quantitative measure of similarity between a pair of rigid 3-D chemical structures.
- Does not require alignment
- Fast (100's a second)
- Different types of atoms can be weighted* An inter-atomic distance matrix is generated for each of the two molecules A and B to be compared
* A Tanmoto similarity is calculated between every atom in molecule A and every atom in molecule B:
similarity = C / NA + NB - C
where C is the number of inter-atomic distances in common (within a margin of 0.5 Angstrom) between the atom in A and the atom in B, and NA and NB are the number of atoms in A and B respectively
* An inter-atomic similarity matix is then used match each atom in A to the most similar atom in B, and the overall inter-molecular similarity is the mean of the similarities of these pairs of atoms
Atom mapping can be weighted by:
Elemental TypesFor more information, see:
Hydrogen-Bonding Classes
Partial Charge Classes
Multivariate Property ClassesPepperrell, C.A., Taylor, R., Willett, P. Implementation and Use of an Atom-Mapping Procedure for Similarity Searching in Databases of Three-Dimensional Chemical Structures, Tetrahedron Computer Methodology, 1990, Vol 3, pp 55-63
2D analysis with VisualiSAR (Example 1 below)View top 200 hits3D analysis with SYBYL (Example 2 below)
Cluster hitsAlign molecules based on mapped atoms (all or just attachment points)
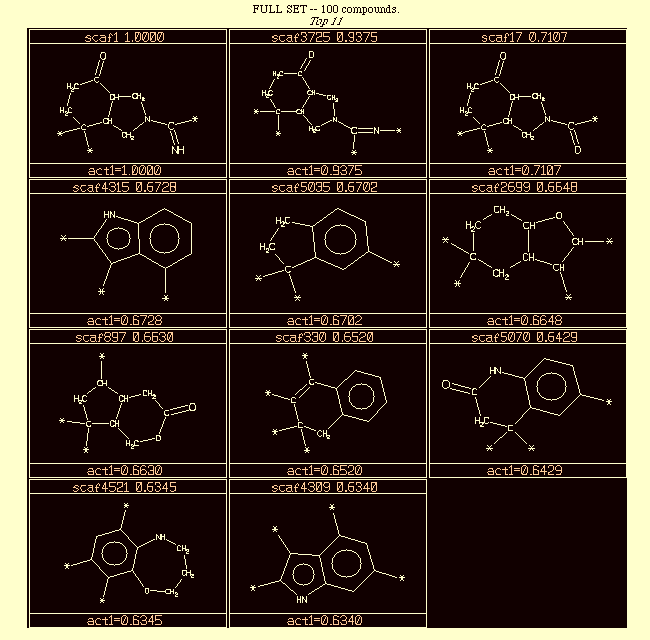
A database of around 6,000 ring systems (scaffolds) was created from the MDDR database.A SAM search was done using Scaf1 (the top left structure in the diagram below) as the query scaffold.
Attachment points were given a weight of 100x other atoms, biasing the search towards scaffolds that have a good overlay of attachment points in three dimensions.
Results from a search of the MDDR are ordered by similarity to scaf1 in VisualiSAR. (e.g. scaf3725 is 0.9375 similar to scaf1) Substituent points are indicated with "*"
The MDDR scaffold database was again usedThis time, Furocinnoline (see below) was used as the scaffold
Attachment points were given a weight of 100x other atoms
Here we look at the top four hits in 3D. For reference, their 2D structures are given below.
Here are the top 4 hits aligned in SYBYL with the query. Purple
atoms indicate scaffold attachment points
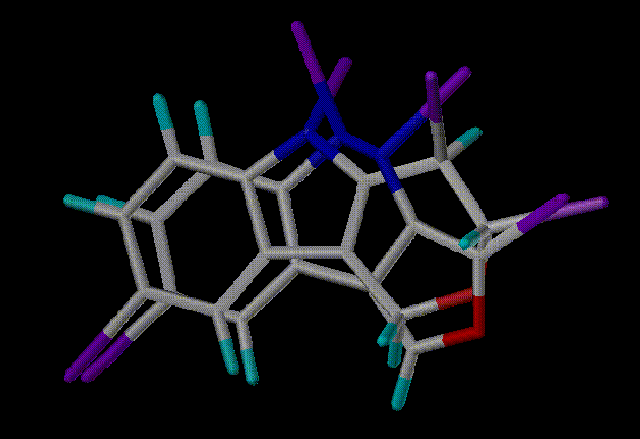 Hit 1 - Scaf2730 - Similarity 0.6687
Hit 1 - Scaf2730 - Similarity 0.6687
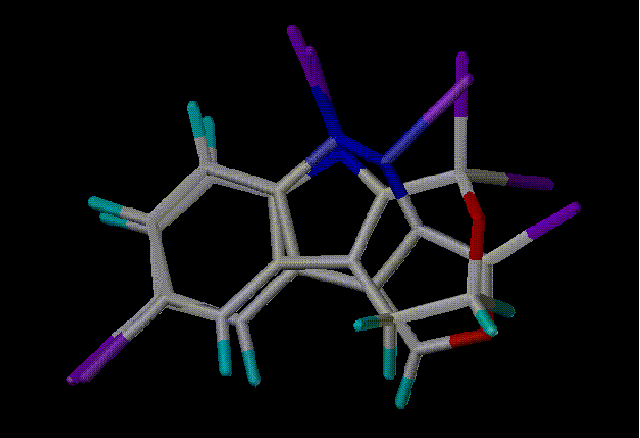 Hit 2 - Scaf 5349 - Similarity 0.6611
Hit 2 - Scaf 5349 - Similarity 0.6611
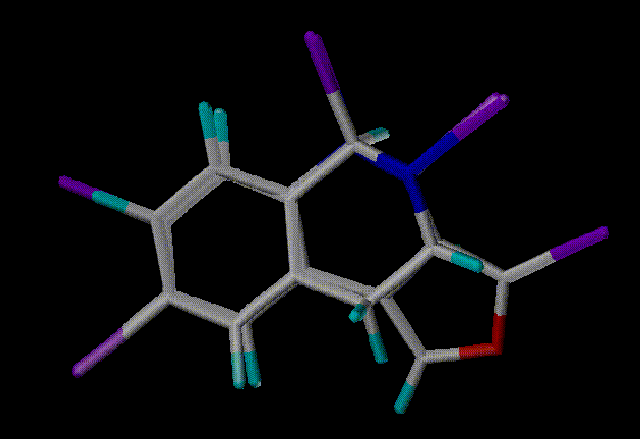 Hit 3 - Scaf 2435 - Similarity 0.6491
Hit 3 - Scaf 2435 - Similarity 0.6491
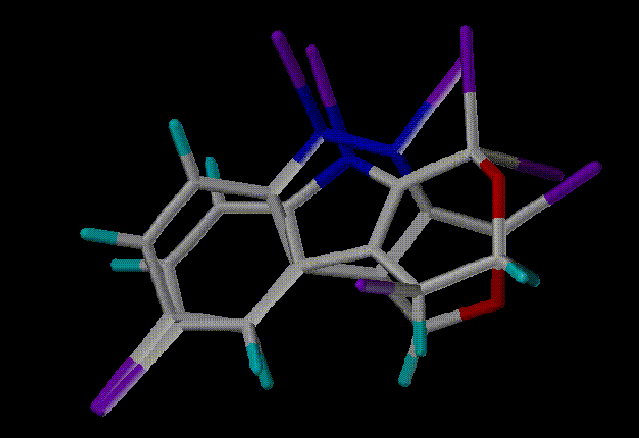 Hit 4 - Scaf 2116 - Similarity 0.6489
Hit 4 - Scaf 2116 - Similarity 0.6489
Finds scaffolds that are clearly not analogs of the query scaffold but which position sidechains in similar positions to the query and have good overal structural similarityFast - can search 6,000-structure MDDR database in around 2 seconds
A good use for some old techniques and new ones!
Highlighting scaffolds / ring systems in clusters using BCI ring fragment dictionaries and Stigmata...
Here the BCI ESSR Ring Fragments were generated for the cluster of penicillins then colored structures were produced in VisualiSAR using the BCI Toolkit
John Blankley, Alain Calvet, George Cowan, Christine Humblet (Parke-Davis)
Peter Willett, Robin & Anne Taylor for permission to use atom-mapping software developed at Zeneca Agrochemicals / Sheffield University