
$DY_ROOT/data/datatypes/std_datatypes.dcis.tdt).
Example Datatype Definition for $SMI
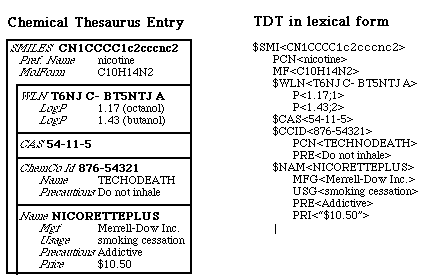
$D<$SMI> _V<SMILES> _B<SMILES> _N<USMILES AUTOGEN $GRF> _P<*> _S<SMILES, primary identifier in the Daylight system> _D<SMILES is the primary key to all data in THOR system. See the Daylight CIS Theory Manual for a complete description.> _M<IDENTIFIER,SYSTEM,POOL> _C<SMILES,the fundamental identifier in THOR databases> _O<Daylight CIS, Inc.> |
| $D<tag> | Defines the internal tag of a new datatype. |
| _V<vtag[;vtag...]> | The verbose tags. This datatype is the only required part of a datatype definition. It serves two purposes: First, it defines the number of datafields in the datatype being defined. Second, it provides the "verbose tags" (human-readable labels) for the datafields of the datatype being defined. |
| _B<btag[;btag...] | The brief tags. A short name for the datatype suitable for labelling buttons and putting in "pull-down menus". |
| _N<ntype[;ntype...]> | The normalizations of each datafield in the datatype. |
| _P<[*][;[*]...]> | The Merlin-pool-inclusion flag. Each non-zero-length field is loaded into Merlin's in-memory pool when Merlin opens the pool. The _P<!> inclusion flag creates a row in Merlin from each subtree rooted by the identifier datatype to which it applies. For example, if set this flag in the $NAM datatype definition, Merlin would create a row for the $SMI(which is standard), but then it would also create a row for each $NAM. |
| _S<summary> | One-line summary of the datatype's meaning and use. |
| _D<description> | Long description of the datatype's meaning and use. |
| _M<set> | Set membership of the datatype. For administration. |
| _C | Comment. You can put anything you like in this datatype. |
| _O | The "owner" of the datatype. You can put anything you like in this
datatype;
|
| $SMI<OCC>2D<1,2,3,4,5,6>| | (original SMILES) |
| $SMI<CCO>2D<5,6,3,4,1,2>| | (unique SMILES) |
For example, one might have two datatypes, NAM and $NAM, the former with just the "AUTOGEN $NAME" and the latter with WHITE0, UPCASE, NOPUNCT. If we entered the dataitem NAM<1,2-dimethylgoo>, a new dataitem, $NAM<12DIMETHYLGOO> would be automatically created.
For example, consider a database for which the ISM<> datatype defined with the "MAKERXNMOL $RMOL,$AMOL,$PMOL" normalization and the three datatypes: $RMOL<>, $AMOL<>, $PMOL<>, each has the USMILES_ANY normalization. If the following datatree is entered:
$SMI<"BrCC=C>>ICC=C"> $RNO<12345> ISM<"BrCC=C>CC(=O)C.CCC(=O)C>ICC=C"> |
the datatree actually stored in Thor, after normalization, would be as follows:
$SMI<"BrCC=C>>ICC=C"> $RNO<12345> ISM<"BrCC=C>CC(=O)C.CCC(=O)C>ICC=C"> $RMOL<BrCC=C> $AMOL<CC(=O)C> $AMOL<CCC(=O)C> $PMOL<ICC=C> |
A molecule's graph is created by removing all isotopic, charge, and bond information from it. All bonds are set to "single", all charges are set to zero, and each atom's hydrogen count is set to the normal lowest valence consistent with its bond configuration. Having removed all of this information, the resulting "molecule" is used to generate a unique SMILES; this is the graph's identifier.
EXAMPLE:
spresi95demo_datatypes.tdt:
spresi95.tdt:$D<"$ISC"> _V<"indirect/citation;citation"> _B<"$ISC/id;$ISC/data"> _P<> _S<Indirect reference for citation data, used in Spresi datatype JA> _M<Indirect> _C<Spresi datatype> _O<Daylight CIS Inc.> | $D<"JA"> _V<"Journal article;Author(s);Institution;Citation;Keywords;Language;Year;Document ID"> _B<"article;art/author;art/inst;art/citation;art/keywords;art/lang;art/yr;art/DocID"> _P<"*;*;*;*;*;*;*;*"> _S<Data type describing a journal article (subclassed from Spresi DOK type).A> _M<Reference> _C<Spresi datatype> _O<Daylight CIS Inc.> |
spresi95_indirect.tdt:$SMI<Br.CCCN(CCC)C1CCc2cc(Cl)c(O)cc2C1> CL<26439;19;;0.0066> FP<e4.02.JEEdR5dQk2EZB.e.YU8Gl6DW326Okrv.uEYE43,AAEN.AX01YU3X2e2Ve7E6Iw45.O0177xQ7YInY34...1;2048;194;512;173;1;S95> TS<199701100502.52> $GRF<Br.CCCN(CCC)C1CCC2CC(Cl)C(O)CC2C1> $SNO<1402584-201> JA<130505;111512;;134485;5~3297~2;ENG;1986;06X0203 87> SMPlt;227.00;227.00;;;;;methanol/diethyl ether;;06X0203 87> |
$ISC<134485;J. MED. CHEM., 29,(1986) N 9, 1615-1627> |
 Daylight Chemical Information Systems Inc.
Daylight Chemical Information Systems Inc.