The distance-as-substructure metric is:
Tversky similariy compares features in a given structure (the "prototype") to features in database structures (as "variants") with user specified weighting for each set of features.
TS = BC / ( BU1 + BU1 + 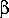 BU2 + BC)
BU2 + BC) |
Example: Setting the weighting of prototype features to 100%
and variant features to 100%, i.e. =1,
=1,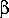 =1,
produces a symmetrical similarity metric identical to the Tanimoto metric.
=1,
produces a symmetrical similarity metric identical to the Tanimoto metric.
Example: Setting the weighting of prototype and variant features
asymmetrically produces a similarity metric in a more-substructural or
more-superstructural sense. Setting the weighting of prototype features
to 100% ( =1) and variant features to 0% (
=1) and variant features to 0% (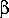 =0)
means that only the prototype features are important, i.e., this produces
a "superstucture-likeness" metric. In this case, a Tversky similarity value
of 1.0 means that all prototype features are represented in the variant,
0.0 that none are.
=0)
means that only the prototype features are important, i.e., this produces
a "superstucture-likeness" metric. In this case, a Tversky similarity value
of 1.0 means that all prototype features are represented in the variant,
0.0 that none are.
Example: Setting the weights to 0% prototype ( =0)
/ 100% variant (
=0)
/ 100% variant (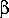 =1) produces a
"substucture-likeness" metric, where completely embedded structures have
a 1.0 value and "near-substructures" have values near 1.0.
=1) produces a
"substucture-likeness" metric, where completely embedded structures have
a 1.0 value and "near-substructures" have values near 1.0.
Tversky metrics where the two weightings add up to 100% (1.0) are of special interest (e.g., the 50/50 metric is known as the Dice index).
 Daylight Chemical Information Systems Inc.
Daylight Chemical Information Systems Inc.