Daylight Fingerprints-
The fingerprint program is included in the Database Package ,Cluster
Package and there is a Fingerprint Toolkit.
Fingerprints were devised to enable high-speed structural screening.
Daylight MolecularFingerprints contain:
-
a pattern for each atom
-
a pattern representing each atom and its nearest neighbors (plus the bonds
that join them)
-
a pattern representing each group of atoms and bonds connected by paths
up to 2 bonds long
-
... atoms and bonds connected by paths up to 3 bonds long
-
... continuing, with paths up to 4, 5, 6, and 7 bonds long. Default is
7, can be up to a max of 31
Example:
the molecule OC=CN
would generate the following patterns:
| 0-bond paths: |
C |
O |
N |
| 1-bond paths: |
OC |
C=C |
CN |
| 2-bond paths: |
OC=C |
C=CN |
| 3-bond paths: |
OC=CN |
-
each pattern sets a set of bits (typically 4 or 5 bits per pattern) which
is added to the fingerprint.
-
If a pattern is a substructure of a molecule, every bit that is
set in the pattern's fingerprint will be set in the molecule's fingerprint.
-
Fingerprints can be variable length (folded) to increase the information
density and decrease the size, to save on storage without creating false
negative results-Fingerprint Density
Reaction Fingerprints
-
Structural Reaction Fingerprints-For Superstructural Matching
-
the fingerprint of the reactant part
-
the fingerprint of the product part
-
the bit-shifted fingerprint of the product part
-
Difference Fingerprints- Reflects bond changes in a Reaction
-
count of each path in the reactant
-
count of each path in the product
-
subtract the counts of a given path
-
if >< 0, then a bit is set in the difference fingerprint
-
if == 0, then no bit is set in the difference fingerprint
Example
Sn2 displacement reaction:
[I-].[Na+].C=CCBr>>[Na+].[Br-].C=CCI
The paths generated for the molecules would be as follows:
| Enumerated Fingerprint Paths: |
| Path Length: |
Reactant (count/path): |
Product (count/path): |
| 0 |
1 I, 1 Na, 3 C, 1 Br |
1 I, 1 Na, 3 C, 1 Br |
| 1 |
1 C=C, 1 C-C, 1 C-Br |
1 C=C, 1 C-C, 1 C-I |
| 2 |
1 C=C-C, 1 C-C-Br |
1 C=C-C, 1 C-C-I |
| 3 |
1 C=C-C-Br |
1 C=C-C-I |
Difference in Path Counts:
|
Path Length:
|
Difference (count/path):
|
0
|
0 I, 0 Na, 0 C, 0 Br
|
1
|
0 C=C, 0 C-C, 1 C-Br, 1 C-I
|
2
|
0 C=C-C, 1 C-C-Br, 1 C-C-I
|
3
|
1 C=C-C-Br, 1 C=C-C-I
|
After generating the difference in counts, we only use the
six paths with non-zero differences to set bits in the difference fingerprint.
These are the paths which walk through bonds that change during the reaction.
By considering only these paths, we get a fingerprint which reflects the
overall bond changes in the reaction.
Mixture Fingerprints-Fingerprint tuples
-
Mixtures stored as Dot Disconnected SMILES are fingerprinted
-
Each component is fingerprinted
-
FPP datatype contains fingerprint for the resulting combination fingerprint
Example:
$SMI<CCC(C)C(N)C(=O)NCC(=O)NC(CO)C(=O)O.CCC(C)C(N)C(=O)NCC(=O)NC(CCCCN)C(=O)O.
CCC(C)C(N)C(=O)NCC(=O)NC(CCSC)C(=O)O.CCC(C)C(N)C(=O)NCC(=O)NC(CC(C)C)C(=O)O.
CCC(C)C(N)C(=O)NCC(=O)NC(Cc1c[nH]cn1)C(=O)O....> FPP<63,59,60,58,56,61,57,62,7,3,4,2,0,5,1,6,39,35,36,34,32,37,33,38,15,11,12,
10,8,13,9,47,43,44,42,40,45,41,55,51,52,50,48,53,49,14,46,54,23,19,20,18,16,21,17,31,27,28,26,24,29,25
,22,30;....E..kcb6Aoe6aF87,0,rr68W,EW0Y.aVYC0J8UQAAedM7.67,VSA,6f.N,FEInJ0Q6ZmUiNZo4kmHJCM0.,CI6...>
$D<FPP>
_V<"Component FP indicies;Component fingerprints;FPP ID">
_B<"FPP/nos;FPP/fps;FPP/id"gt; _N<"PART_NTUPLE 1;BINARY;">
_P<"*;*;"gt; _S<"Component fingerprint indicies;Component
fingerprints;FPP ID">
_M<System>
_O<Daylight Chemical Information Systems Inc.>
Fingerprint options:
-
Minimum/Maximum Size (power of 2, typically 1024 for small molecules)
-
Density (0.3)
-
Minimum/Maximum Path length
-
Difference Fingerprints
-
Mixture Fingerprints
Comparing Fingerprints-
Three Similarity Metrics
-
Tanimoto Coefficient
-
Euclidian Distance
-
Tversky Similarity
| Symbol |
Definition |
Description |
| bits(F) |
|
A function that returns the number of "1" bits in a bitmap |
| BT |
|
The total number of bits (the fingerprint's size); a constant |
| B1 = |
bits(F1) |
The number of 1's in F1 |
| B2 = |
bits(F2) |
The number of 1's in F2 |
| BC = |
bits( F1 AND F2 ) |
The number of 1's in common between F1 and F2 |
| BI = |
bits(F1 XOR (NOT F2)) |
The number of identical bits (1's and 0's) between F1 and F2 |
| BU1 = |
bits(F1 AND (NOT F2)) |
The number of unique bits (1's) in F1 |
| BU2 = |
bits(F2 AND (NOT F1)) |
The number of unique bits (1's) in F2 |
Tanimoto Coefficient- the number of bits in common divided
by the total number of bits that could be in common. Scale, 1.0
identical fingerprints, 0.7 highly similar, 0.5 roughly similar
Euclidian distance- a measure of the geometric distance between
two fingerprints. Scale , 0.0 identical fingerprints, 0.3 highly similar,
0.5 roughly similar
| DE(F1,F2) = (BT - BI) / BT |
The distance-as-substructure metric is:
| DSE(F1,F2) = (B1 - bits(F1 AND F2)) / B1 |
Tversky Similarity-
For a complete description of Tversky similarity see John Bradshaw's
MUG '97 presentation, "Introduction
to Tversky similarity measure".
-
Tversky similariy compares features in a given structure (the "prototype")
to features in database structures (as "variants") with user specified
weighting for each set of features.
TS = BC / (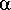 BU1 + BU1 + 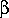 BU2 + BC)
BU2 + BC) |
Example, Setting the weighting of prototype features to 100%
and variant features to 100%, i.e. 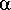 =1,
=1, 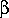 =1,
produces a symmetrical similarity metric identical to the Tanimoto metric.
=1,
produces a symmetrical similarity metric identical to the Tanimoto metric.
Example, Setting the weighting of prototype and variant features
asymmetrically produces a similarity metric in a more-substructural or
more-superstructural sense. Setting the weighting of prototype features
to 100% (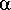 =1) and variant features to 0% (
=1) and variant features to 0% (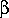 =0)
means that only the prototype features are important, i.e., this produces
a "superstucture-likeness" metric. In this case, a Tversky similarity value
of 1.0 means that all prototype features are represented in the variant,
0.0 that none are.
=0)
means that only the prototype features are important, i.e., this produces
a "superstucture-likeness" metric. In this case, a Tversky similarity value
of 1.0 means that all prototype features are represented in the variant,
0.0 that none are.
Example, etting the weights to 0% prototype (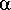 =0)
/ 100% variant (
=0)
/ 100% variant (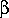 =1) produces a
"substucture-likeness" metric, where completely embedded structures have
a 1.0 value and "near-substructures" have values near 1.0.
=1) produces a
"substucture-likeness" metric, where completely embedded structures have
a 1.0 value and "near-substructures" have values near 1.0.
-
Tversky metrics where the two weightings add up to 100% (1.0) are
of special interest (e.g., the 50/50 metric is known as the Dice index).
 Daylight Chemical Information Systems Inc.
Daylight Chemical Information Systems Inc.
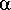 BU1 +
BU1 + 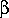 BU2 + BC)
BU2 + BC)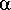 BU1 +
BU1 + 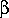 BU2 + BC)
BU2 + BC)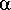 =1,
=1, 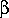 =1,
produces a symmetrical similarity metric identical to the Tanimoto metric.
=1,
produces a symmetrical similarity metric identical to the Tanimoto metric.
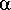 =1) and variant features to 0% (
=1) and variant features to 0% (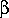 =0)
means that only the prototype features are important, i.e., this produces
a "superstucture-likeness" metric. In this case, a Tversky similarity value
of 1.0 means that all prototype features are represented in the variant,
0.0 that none are.
=0)
means that only the prototype features are important, i.e., this produces
a "superstucture-likeness" metric. In this case, a Tversky similarity value
of 1.0 means that all prototype features are represented in the variant,
0.0 that none are.
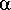 =0)
/ 100% variant (
=0)
/ 100% variant (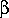 =1) produces a
"substucture-likeness" metric, where completely embedded structures have
a 1.0 value and "near-substructures" have values near 1.0.
=1) produces a
"substucture-likeness" metric, where completely embedded structures have
a 1.0 value and "near-substructures" have values near 1.0.
 Daylight Chemical Information Systems Inc.
Daylight Chemical Information Systems Inc.