The Hitch Hiker’s Guide
to Chemical Space
by Anthony Nicholls,
OpenEye Scientific Software, Inc.
![]()
From the Introduction to "The Hitch Hiker’s Guide to Chemical Space":
Q:What is Chemical Space?
A: It's where 10200 small, possibly drug-like, molecules live.
Q: 10200. Isn’t that a big number?
A: Yes, it's big. Really big. You just won't believe how vastly hugely mindbogglingly big it is. I mean, you think your average combinatorial library is big, but that's just peanuts compared to Chemical Space. Listen:
(it goes on for a bit here before finally settling down with things you really need to know)
Q. Sounds scary. Do you have anything helpful to say about finding your way around in Chemical Space?
A: Lots! For starters…

![]()
The important Question:
In a
10200, how many are really different?
Only differences in:
that produce differences in:
really matter (for drug design).
![]()
Fundamental OpenEye Credo:
Shape Matters
Shape = Field Properties:
Such as:
Steric
Electrostatic
Functions derived by atoms types
(e.g. hydrophobic, hydrogen bond potential)
![]()
Why we feel shape is probably sufficient
Consider the following problems in traditional energetic approaches to the important problem of calculating binding energies:
i.e. why worry about "precise" vacuum energies?!?
![]()
To have a shape one needs a structure:
Computational Structure Generation
General Methods:
However, structure generation has to take account of the aqueous environment of biological processes, i.e. we need:
Solvation Modeling:
The output from Solvent Modeling has two uses:
Possible Methods include:
We use PB because we believe it provides the highest ratio of accuracy to CPU cycles, once certain implementation problems are addressed.
![]()
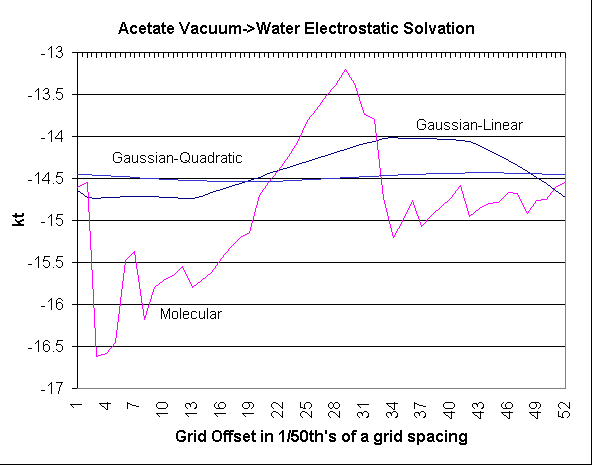
OpenEye Advances in PB
Results:
![]()
Comparing Molecular Shapes
(i.e. what is the overlap of A with B)
(e.g. think of a grid/lattice representation)
(admittedly in an infinite dimensional space)
(aka: the triangle inequality: dab + dbc >= dac rules, ok)
![]()
Why are metrics so wonderful?
Example 1:
Exhaustive Search for the Best
Overlay = Minimal Field Difference
Traditionally a 6-dimensional search problem
But Consider:
Example 2:
Minimal Metrics =
Metric of a Different Order
Given a non-projective Operation T
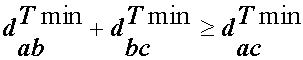
i.e. the mimimum field difference between two molecules is also a metric.
An example of T would be a rotation and/or translation, and hence the
best overlay between two molecules defines an interesting minimal metric, in particular for the space it "induces".![]()
Shape Space
The N*(N-1)/2 optimal overlays of N molecular fields form an N*N distance matrix,
D.
From a distance matrix can be derived what is known as the metric
G matrix. If G is diagonalized the number of approximately non-zero eigenvalues determines the "effective" dimensionality of the geometric space N points can inhabit to give rise to those N*(N-1)/2 distances, and the position of those points.
The space formed from the diagonalization, and subsequent culling of insignificant dimensions, of the metric matrix derived from a set of molecular overlay distances we refer to as
Shape Space.
Q: What is the dimensionality of the Shape Space of
10200 small molecules?A: Probably that of a MUCH smaller set
Q. How much smaller?
10180, 1020 (if so we’re in trouble!) or 102 ?
We plan to find out via the fields of the structures corresponding to:
My Guess? Dimensionality < 100. Why? Because, as we shall show in later, the steric field can usually be approximated very well by 30-40 variables. We anticipate a smaller dimensionality for electrostatic fields.
Uses of a Shape Space Decomposition:
PLUS: Tames 10200 molecules, sort of.
![]()
All of the above concerns global similarity, but what of:
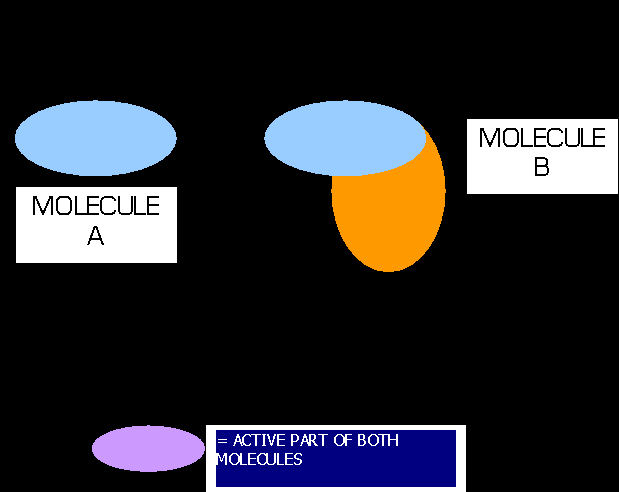
i.e. A and B fit poorly on top of each other, and yet they both have an essential element of common shape.
![]()
Local Shape Comparisons:
Two Novel Metric Approaches:
Ellipsoidal Domain Decomposition
The Idea:
Use a smooth gaussian function for the molecular field (typically steric) and seed N ellipsoidal gaussians within the molecules. Minimize the variable describing these gaussians against the field difference from the sum of such and the molecular field
The Result:
(for 1-3 Ellipsoidal Gaussians for a small, 2 ringed molecule)
One Ellipsoid Fit
Two Ellipsoid Fit
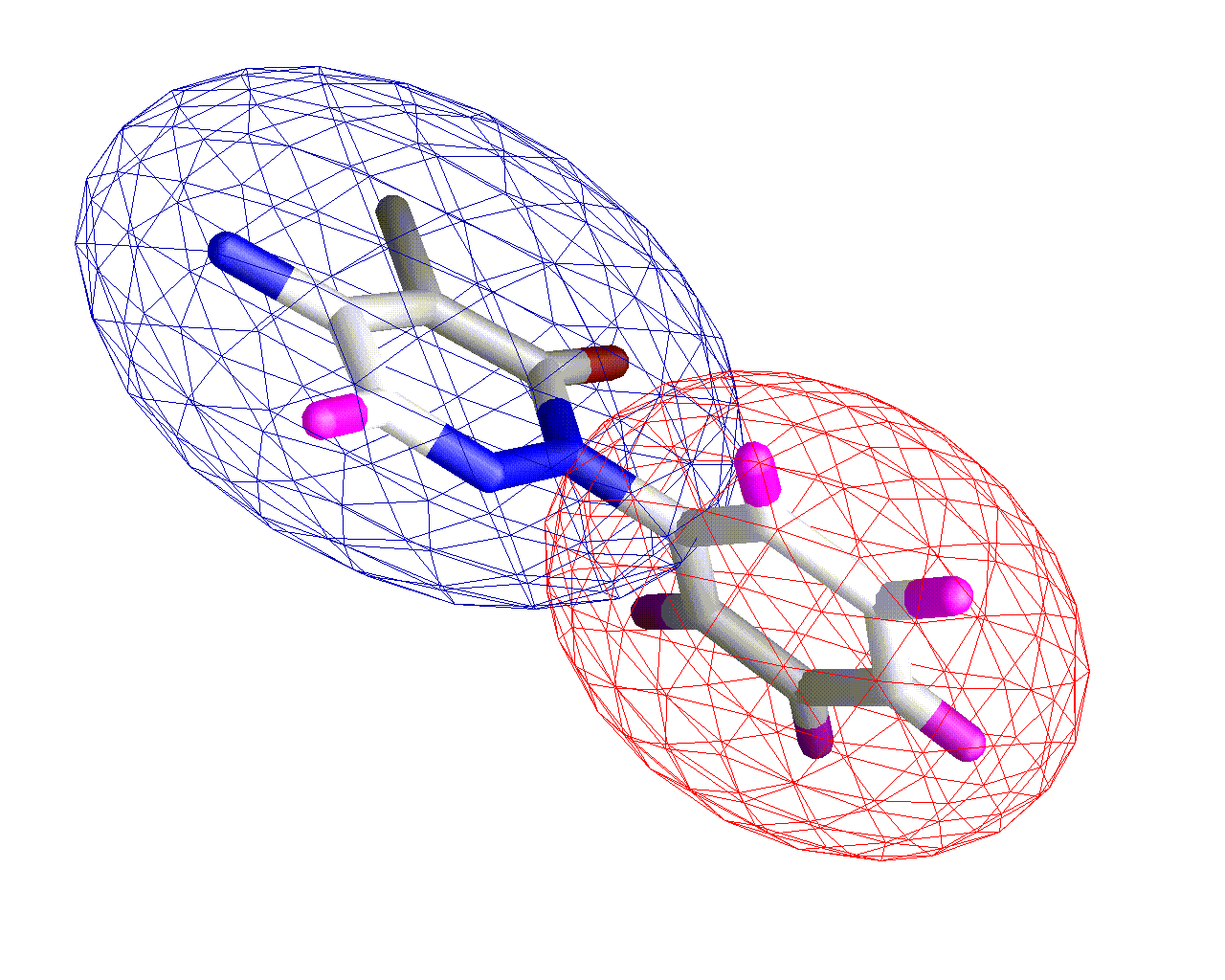
Three Ellipsoid Fit
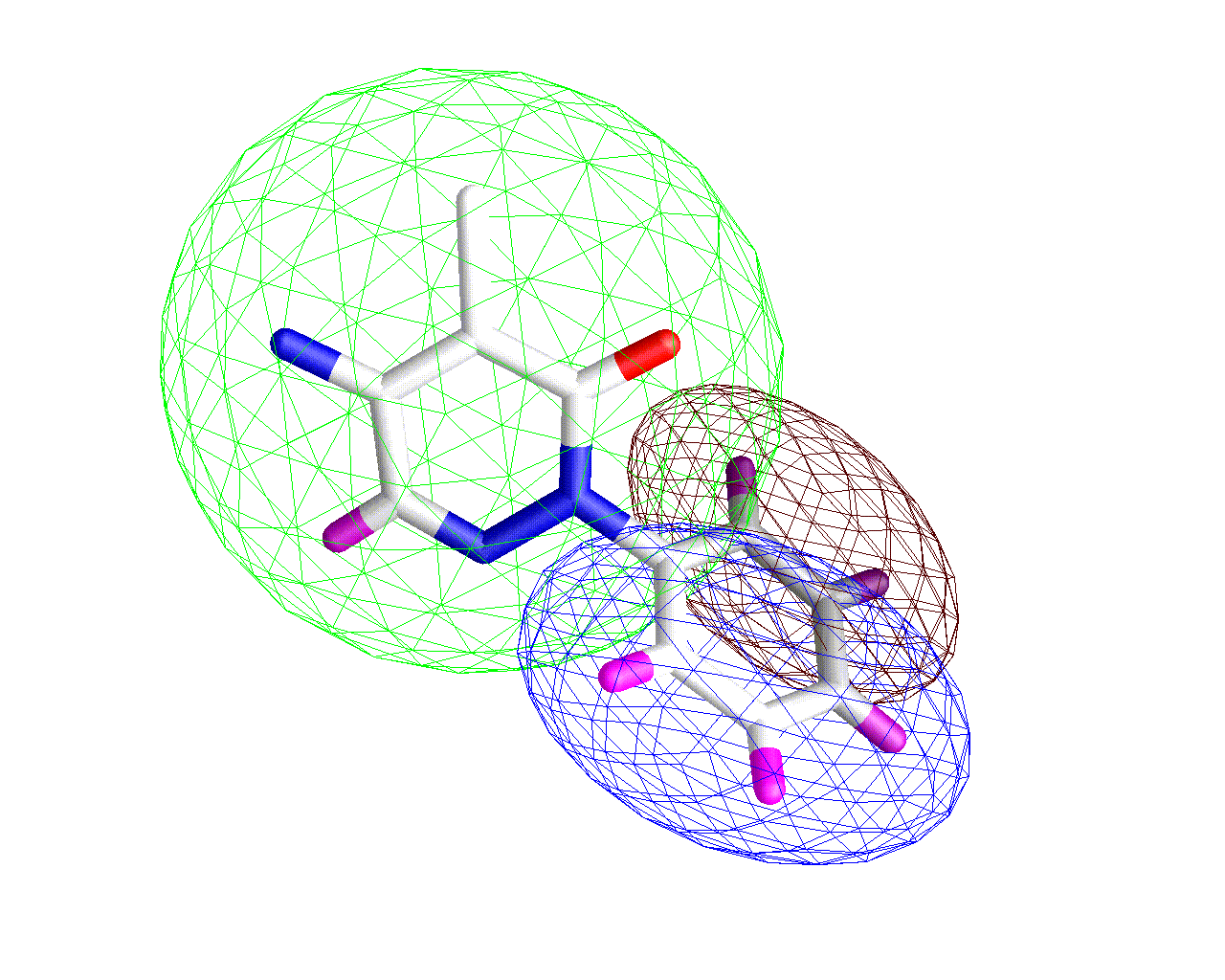
By analyzing the fit of each ellipsoid to the underlying atoms we can robustly determine that the 2 ellipsoid fit is the best representation for this molecule.
Most molecules are fit by 3-4 ellipsoids to within 10-15A3, i.e. less than volume of one methyl group. As each ellipsoid has 10 free parameters, the steric field can thus be well represented by 30-40 parameters, and hence our hope for a shape space of similar dimensionality.
![]()
Uses of Ellipsoidal Decompositions:
![]()
Final Conclusions:
"What looks like molecule X?":
![]()
Finally, Like Hitch Hiking, Waiting for OpenEye Tools requires Patience!
Current State of OpenEye Code:
Until then:
KEEP
BANGING
THE
ROCKS
TOGETHER!
![]()
Thanks to:
Peter Jeffs @GlaxoWellcome and Tony Wilkinson @Zeneca for funding for some of these projects
Andy Grant @Zeneca for his work, ideas and enthusiasm
Roger Sayle for help and wise thoughts
Daylight for the chance to present
Dave-I-Think-Big-Therefore-I-Am-Weininger for the courage to try