Assembling off-the-shelf Components Into Useful Applications,
TJ O'Donnell, MUG2004
parse_smiles
This is a set of Perl functions which parse SMILES/SMARTS strings.
They are useful for modifying SMILES/SMARTS
output by various drawing applications. For example, one might
correct systematic errors, convert to standard Daylight
SMILES/SMARTS, insert atoms, etc.
Details:
- @aindex = &index_atoms($smi);
index_atoms parses a smiles and returns an array indicating the string location and length of each atom symbol.
- @atoms = &get_atoms ($smi, \@aindex);
get_atoms returns an array containing the strings which are the atom symbols in the input smiles;
@aindex is the array output by the index_atoms function.
- $newsmi = &make_smiles($smi, \@index, \@atoms);
make_smiles reconstructs a smiles from an input smiles, a atom index (from index_atoms function)
and an array of atom symbols (@atoms).
The sequence of function calls, as shown above, would produce a $newsmi
identical to the input $smi. However, it is possible to change
the @atoms array after &get_atoms and before &make_smiles. This
would result in a $newsmi with changed atoms symbols, but all other
ring and bond symbols unchanged. Current uses of parse_smiles is
in the
SAR
application to insert [R1], [R2], etc. into a smiles to indicate
where substitutions have been detected.
Future uses (and possible additional functions) will allow one
to temporarily change a SMARTS into a valid SMILES in order to process the
SMARTS using all the dt_ functions which normally operate only
on valid SMILES. This would enable things like saturate a
SMARTS with H atoms, for example:
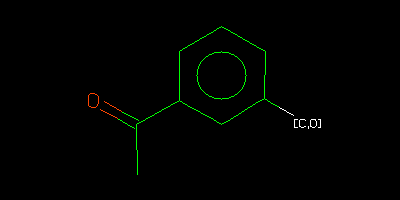 c1c([C,O])cccc1C(=O)C
c1c([C,O])cccc1C(=O)C
could become
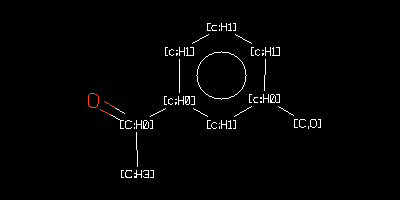 [c;H1]1[c;H0]([C,O])[c;H1][c;H1][c;H1][c;H0]1[C;H0](=O)[C;H3]
[c;H1]1[c;H0]([C,O])[c;H1][c;H1][c;H1][c;H0]1[C;H0](=O)[C;H3]
 c1c([C,O])cccc1C(=O)C
c1c([C,O])cccc1C(=O)C
 [c;H1]1[c;H0]([C,O])[c;H1][c;H1][c;H1][c;H0]1[C;H0](=O)[C;H3]
[c;H1]1[c;H0]([C,O])[c;H1][c;H1][c;H1][c;H0]1[C;H0](=O)[C;H3]