The Senomyx
Discovery Process
Senomyx
Mission:
To discover new and improved flavor and fragrance molecules through integrating our enhanced understanding of the biology of taste and smell with high throughput molecular discovery technology.
High Throughput Sensory Discovery:
A
rapid, iterative process based on a system of tightly integrated and automated
just-in-time components of molecular discovery discipline.
• Knowledge-based Computational Library Design
• High Throughput Synthesis
• Analysis and Purification
• Assay Development
• High Throughput Screening
Informatics
Foundation and infrastructure:
• Oracle Database (Senobase)
• Daylight Oracle Cartridge and toolkits
• Modular server-side software components (VB, Java)
• Web-based user interfaces
• Numerous behind the scenes middleware that sets the foundation
for rapid application development.
Informatics Tools:
·
Structure/substructure
searching
·
List
generation
·
List
management
·
Method
writer
Computational
molecular design:
·
Diversity
space is limited with matrix synthesis
·
Goal
is to expand diversity space through cherry picking
·
Selection
from virtual library
·
2-D
topology-based methods
·
Nearest
neighbor approach
·
Genetic
algorithm used for searching
·
Cherry
picking hundreds for synthesis
·
Fully
integrated into workflow
Virtual
libraries:
·
Defined
by a generic reaction (A+B+C => ABC)
·
Scope
of viable reagents
·
Novelty
and relevance
·
The
sum of all virtual libraries constitutes the search space for target guided
design
Genetic
Algorithm:
1. Start with a randomly generated seed
population of n candidate reactions or from a pre-existing population
2. Repeat the following steps until n offspring have been created:
·
Select
a pair of parent candidates from the current population
·
Apply
a singe-point crossover operator in a chemically meaningful way; keep one new
offspring randomly
·
Mutate
the offspring with specified mutation probability
3. Combine new and old populations
4. Calculate the fitness of each candidate in the combined
population
5. Sort population by fitness and cull the less fit half.
6. Go to step 2.
Fitness based selection:
•Maximize
activity in biological assay
Use a “training set” of compounds whose activities have been measured in a biological assay. Estimate the activity of each new compound by selecting its 10 nearest neighbors from the training set and taking a weighted average of their activities. Nearest neighbors are determined by Tanimoto coefficients of similarity for 2D binary fingerprint comparison.
•Maximize
population diversity
Diversity of a given set of compounds D(S) is estimated as
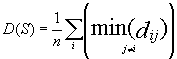
where n is the number of compounds in S and d
is the diversity between two
molecules based on Tanimoto coefficient for binary
fingerprint comparison
Ref. Dimitris K. Agrafiotis & Victor S. Lobanov, J.
Chem. Inf. Comput. Sci. 1999, 39, 51-58
•Optimize
reagent use
Automated
library synthesis:
·
Chemistry
definition (reaction transform – SMIRKS
– and building block lists)
·
Library
enumeration (GA)
·
Run
preparation (material balance)
·
Chemistry
services (bulk to automation ready)
·
Automated
synthesis
·
Plate
reformatting
·
Purification
and LC/MS analysis
·
High
throughput screening
Summary:
• Design,
synthesis, and screening cycle is ~1 week
• Current
throughput is several thousand compounds per week
• >50,000
compounds have been synthesized and screened using more than 40 automated
combinatorial reactions
• Hundreds of
active molecules have been identified from a total of over 10 chemical classes