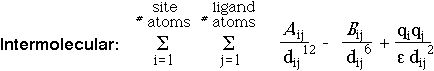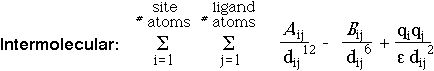Dockit DG dock engine
-
After the ligand is inside all site spheres, the spheres are removed and conjugate gradient minimization continues in 4 dimensions vs. the distance geometry intramolecular error function and an intermolecular steric and electrostatic function:
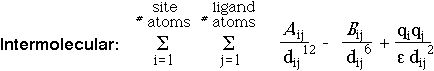
-
United atoms with no explicit hydrogens are used; charges are assigned by analogy to AMBER using SMARTS rule-based assignment.
-
Docking is scored initially by the final, optimized intermolecular energy.