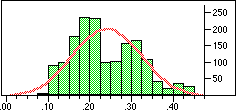
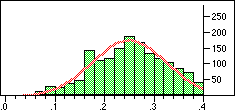
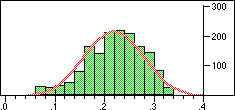
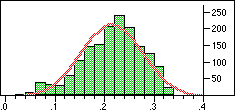
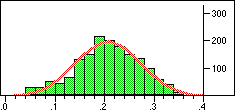
| Distribution of Tanimoto scores for folded fingerprints against vitamin a |
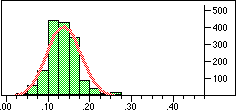
Distribution of Tanimoto scores for non-folded fingerprints against vitamin a |
|---|---|
 |
 |
John Bradshaw and
Roger A. Sayle
Glaxo Wellcome, Stevenage, Herts SG1 2NY, UK
Two of the FAQ about molecular similarity and diversity are
Alternatively, as we hold the view that the data structure should be determined by the data and not by some arbitrary imposed cartesian grid, we can attempt to add some statistics which are set and target based in order to address these problems.
By borrowing the methods used in bioinformatics for similarity scores ( J.F. Collins, A.F.W. Coulson and A.Lyall CABIOS (1988) 4(1) 67-71), we attempt in this paper/poster to indicate possible answers to the two questions posed.
Is the similarity between the object I am considering and the target significantly greater than one would expect by chanceIn a preliminary experiment we looked at the distribution of similarity scores of a set of typical drugs ($DY_ROOT/data/drugs.smi) against the wdi971demo database provided in the 4.51 release of the DAYLIGHT software. If we ignore the nearest 100 neighbours then the distribution of similarity scores is almost normal. Note this is not true if we use folded fingerprints.
or
Is the similarity between the object I am considering and the target significantly greater than the mean value to all other objects in the set
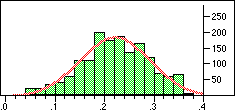
| Distribution of Tanimoto scores for folded fingerprints against vitamin a |
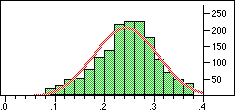
Distribution of Tanimoto scores for non-folded fingerprints against vitamin a |
|---|---|
 |
 |
If we require that significant neighbours are at least 5 standard deviations (i.e. a Z-score > 5 )from the mean of the similarity to the non-immediate neighbour group then we get the table shown below.
| Structure | Distribution of Tanimoto scores | Z-score | Number of significant neighbours Click to view |
|---|---|---|---|
 |
|||
 |
|||
 |
|||
 |
|||
 |
|||
 |
|||
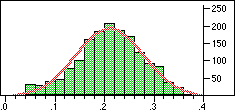 |
|||
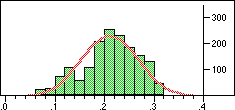 |
|||
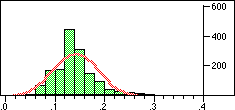 |
|||
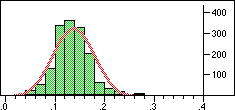 |
Note that in the case of caffeine we have significant neighbours below 0.4 on a Tanimoto scale, illustrating that the significance is set dependent.
Will this new compound usefully increase the diversity of my compound collection?One of the difficulties with the measurement of diversity is that it needs to be linked to the appropriateness of the task. For example if one wished to cross the Atlantic by diverse forms of transport, you would very rapidly eliminate a bicycle from your list of options as it is not appropriate for the task. In the same way, certain compounds are inappropriate as drug lead candidates. However the measure of appropriateness is difficult.
One approach is to order a collection by its Z-score relative to the rest of the database. If you do this then the "odd-ball" compounds rise to the top. For instance, if we calculate the Z-scores for the wdi971demo database and sort the database by the score, the ones with the highest scores would seem inappropriate as drug leads.
However if we look at the compounds with the lowest overall Z-scores, they would seem to be much more appropriate as drug leads.
Initial studies would indicate that a Z-score less than 18 in this database would appear to be reasonable for a drug lead candidate.
Potentially, therefore we have a way of 'cleaning' databases and removing inappropriate compounds without a priori having a rigorous definition of appropriateness.
In a single experiment we took the 1779 compounds in wdi971demo and calculated
the Z-scores against the wdi971demo database and against the spresi95demo databases.
The hope was that we could spread the compounds and get some handle on drug likeness. In the event the values were highly correlated
We also need to estimate how many neighbours we need to skip when calculating the population mean. Currently we cannot use any of the fast methods for means of pairwise similarities, as the whole target set needs to be sorted before the exclusion. Experiments are being carried out excluding compounds which could not possibly be, say, within a Tanimoto similarity of 0.8 as the do not have sufficient bits set. Results are currently inconclusive.
Unfortunately the current code works against a merlin database and as the merlinload uses the *.DP file rather than a dt_stream() across the database, see dt_open(3), there is currently no simple way to produce "random" sample merlin bases from a parent Thor database. (See also Dave's comments for further detail.)
One of us has suggested that the ratio of the Z-scores may provide a useful similarity index. This will require investigation, not least because the resulting index will be directional.
This work would not affect a clustering method such as Jarvis Patrick as the ordering of nearest neighbours to a target is not affected. However hierarchical methods become difficult as the Z-scores are intrinsically directional.