Neural net clustering
Euromug '96
- New projects
- previous
- next
Synopsis
The project goal is to examine neural network based clustering methods
which operate in O(N) time.
If such a method could be developed which produces meaningful clusters,
it would provide a great speed improvement over the best methods
currently used for large databases which are O(N^2).
People
- Jack Delany (wrote prototype)
- Kai Hasselbach (concept and collaboration)
- Johann Gastieger (Kohonen algorithm advocate)
Status
A proof-of-prinicple implementation has been written and subjected to
preliminary analyses. Indications are that this approach to clustering
shows promise for large database applications but will require a
significant development effort to bring to a production level.
Development is on hold pending evaluation of customer interest.
Description
The prototype program is an implementation of Kohonen neural net
algorithm adapted to clustering binary fingerprint data.
A brief description follows with numbers per the prototype
(which are almost certainly not optimal).
A rectangular neural network grid of 20x20 neurons
is set up as a torus to avoid edge effects
(both pairs of opposite edges wrap around)
Each of these 400 neurons is composed
of 256 real values corresponding
to the bits in a fixed length fingerprint.
Neuron values are initialized to
uniform random numbers in the range 0.0 to 1.0.
A training pass consists of training the network to
all 32,469 molecules in the medchem96 database
one at a time, in random order.
For each molecule, the neuron is found which is most similar to the
input molecule using Tanimoto metric of fingerprint similarity.
The values for that neuron are adjusted by a weighted average which
decreases linearly though the pass.
Nearby neurons as far as 4 grid units away also have their
values adjusted with weights decreasing linearly with distance.
A number of passes (3) is required to train such a network.
Once the network is trained, molecules are assigned to the neuron with
the most similar value.
Molecules assigned to the same neuron are considered to be in the
same cluster.
Here is an example of some of the clusters produced using the method
described above:
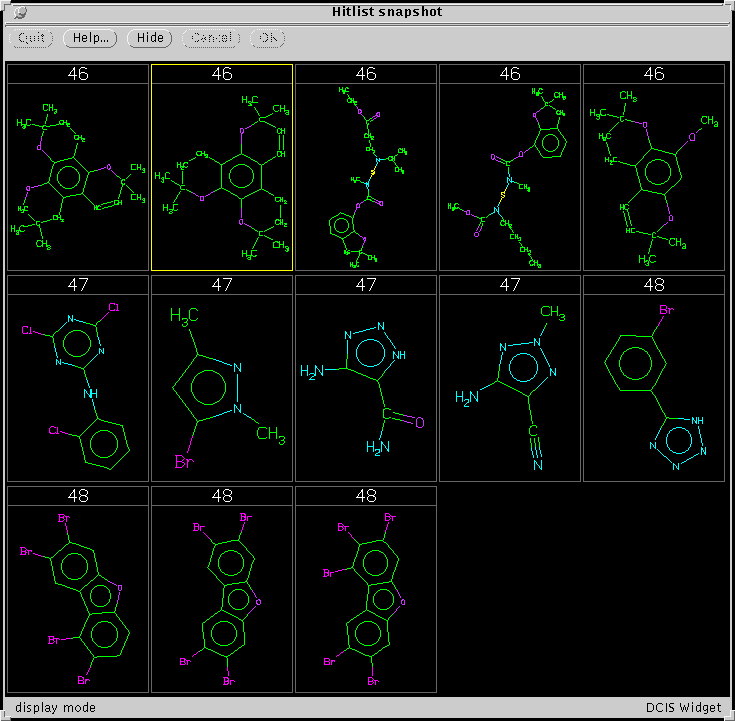
We have not thoroughly analyzed this algorithm,
but initial conclusions are mixed.
The good news is that the program does produce clusters,
it does operate in O(N) time,
and it does produce results which are interpretable by conventional
clustering methods (e.g., dendograms).
The bad news is that
training passes are surprisingly slow (8 CPU-hours/pass),
clusters produced by few (3) training passes are not
particularly intuitive,
and several peculiarities of the method have become apparent
(e.g., order dependency).
It is clear that optimizing this program will require a significant
amount of effort.
We would need to study the effects of various parameters and their
interactions
(e.g., network size,
training and adacency schedules,
effect of randomizing order in different training passes,
etc.)
It is probable that large speed optimizations could be found
although this algorithm is relatively difficult to parallelize.
Before committing to this work, we are seeking feedback from
users as to the perceived value of such a product.
 Daylight Chemical Information Systems, Inc.
Daylight Chemical Information Systems, Inc.
info@daylight.com

