EuroMUG '04 -- 4 - 5 Nov, 2004
Conversion Toolkit & Applications
Michael A. Kappler
Daylight CIS, Inc.
ABSTRACT
A description of the Conversion Tools project and its initial release state is presented. The tools are scheduled to be included in Daylight Software v4.9.
Introduction
Chemical information is stored in a variety of ways.
- Languages
- Formats
- Chemical EXchange
- Thor DataTree
Daylight surveyed the chemical information field to determine the popularity of formats and sketchers. We found a significant portion (80-90%) of chemical information is stored in Molecular Design Limited (MDL) chemical table file (CTfile) formats.
- molfile
- RDfile
- RGfile
- rxnfile
- SDfile
- SData
- extended molfile
- extended rxnfile
Most chemists use either ChemDraw or ISIS-Draw as a sketcher and each has its own format.
Currently, our Open Source (``Contrib'') programs MOL2SMI and SMI2MOL perform conversion to and from CTfile formats.
- molfile and SDfile only
- estimated correct conversion rate of 99.9%
- incorrect conversion of 0.1% of a large database could be hundreds or thousands of structures.
Software, such as OpenBabel, is available to convert between many formats including SMILES.
- however they either fail to properly handle the language
- do not support other Daylight languages
Similarly, ChemDraw and ISIS-Draw generate SMILES.
- but neither do it according to our standards
Goals
The goal for the initial Conversion Toolkit release is to satisfy the majority of:
- our customers needs
- our internal needs
- potential use by future customers
Objectives
We set out to develop a general tool for conversion of chemical information to and from Daylight languages. Realization of this project will manifest in the form of a Daylight Toolkit, a set of applications, and server side functionality in DayCart. Specific objectives include:
- minimizing the number of supported file formats
- leaving the door open for extensibility
- focus on the CTfile formats and the ChemDraw sketcher
Other formats such as XML/CML and sketchers such as ISIS-Draw, MDL-Draw, Marvin and JME will be considered at a later date
Methods
Design. The Conversion Toolkit will be a static and dynamic link library and integrated with existing tools.
- The Toolkit approach maximizes robustness - it should never itself fail, only fail to convert malformed input.
- Users will not be required to know the input format, for example, where a structure ends and another begins within a SDfile.
- Direct I/O will be performed by applications, not the toolkit itself.
- The essence of the Conversion Toolkit will become part of DayCart.
Approach. The Conversion Toolkit interface will accept an object and input data, and outputs data in the correct format.
- The object will define the desired conversion, e.g., convert from a MDL SDfile to Daylight SMILES.
- Also, perception of input data will allow generic conversion of data.
- The interface will be extensible in order to easily support additional formats.
Issues. Dialects increase the complexity of converting a format. Do we take a hard or soft line on violations?
Technical Details
Toolkit. There will be two main routines in the Conversion Toolkit. The first routine creates an object for a particular type of conversion.
dt_Handle <= dt_alloc_conversion(dt_Integer ifmt, dt_Integer ofmt)
- The parameters ifmt and ofmt will indicate the input and output formats, such as SDfile and SMILES.
- There will be integral definitions in the toolkit for format types.
- DX_CONV_FMT_SMI
- DX_CONV_FMT_SMA
- DX_CONV_FMT_SMR
- DX_CONV_FMT_MOL
- DX_CONV_FMT_RDF
- DX_CONV_FMT_RGF
- DX_CONV_FMT_SDF
- DX_CONV_FMT_SDT
- DX_CONV_FMT_V30
- DX_CONV_FMT_V3R
- For example, to indicate a SDfile to SMILES conversion, the input and output parameters will be DX_CONV_FMT_SDF and DX_CONV_FMT_SMI, respectively.
Two powerful generic types will be provided as well.
- The type DX_CONV_FMT_MDL will indicate any of the CTfile formats, and will not dictate which one or require that it be homogeneous. This is useful for converting an input stream of mixed types, such as a SDfile followed by a RDfile, or a RDfile containing molecules and reactions.
- The type DX_CONV_FMT_DAY will indicate any of the Daylight languages and will not dictate which one.
- In the case of generic types, the actual input type will be perceived and the actual output type will vary according to the input type.
For toolkit programmers, the type DX_CONV_FMT_OBJ will indicate a Daylight object, such as a molecule, reaction, pattern, or transformation.
The return value from the dt_alloc_conversion routine will be a conversion object that is passed with data into the second routine, which converts data from one format to another.
dt_Handle <= dt_convert(dt_Handle conv, dt_Handle seq)
The parameter conv will be the conversion object from above and seq will be a sequence of strings or objects. Named Properties associated with the conversion object will control the behavior of the conversion process. Depending on the output format, the return value will be either a sequence of strings or objects. For example, when converting from a SDfile to SMILES, the input and output data will be a sequence of string objects representing a SDfile (one line per string) and SMILES (one structure per string). In the case of conversion to objects, the output data will be a sequence of dt_Handle types (molecules, reactions, etc.).
The list of supported formats will be as follows: MDL CTfile formats except XDfile (molfile, RGfile, rxnfile, SDfile, RDfile, extended molfile and extended rxnfile), SMILES, SMARTS, SMIRKS, Daylight Toolkit objects (molecule, reaction, pattern, and transform), and TDTs. Any CTfile format may be converted to the corresponding Daylight language or object, and vice versa.
Applications. Two powerful generic converters, mdl2daylight and daylight2mdl, will be provided for conversion to and from all of the supported MDL and Daylight formats. These converters perceive the input type and generate the most appropriate output type. The choice of which output type will be produced from an input type will be predetermined. For example, one input SMILES results in a molfile and multiple input SMILES result in a SDfile. The ability to recognize MDL input type will be limited. When the input or output type is known, the format may be specified.
All applications will be built on an internal conversion utility and support streaming of data, also known as \emph{feeder mode}. Conversion of data will be performed in parts and the size of the parts may be set by the user. Feeder mode will be necessary for processing large input files that would otherwise exceed hardware capabilities.
The main programming loop condition is:
while (NULL_OB != (oseq = dt_convert(conv, iseq)))
The parameter iseq will be filled with data prior to calling the dt_convert routine. Within the loop, converted data will be output and additional data will be placed in the input sequence.
Toolkit programmers wishing to convert to Daylight Toolkits objects (molecule, reaction, etc.), need to write their own programming loop to capture the objects as they are returned from \texttt{dt_convert}. For example, pseudo code for conversion of a SDfile to molecules is as follows:
#include "dt_smiles.h" /* SMILES Toolkit */
#include "dt_conv.h" /* Conversion Toolkit */
int main() {
dt_Handle conv, iseq, oseq;
/* create conversion object */
conv = dt_alloc_conversion(DX_CONV_FMT_SDF, DX_CONV_FMT_OBJ);
/* create input object */
iseq = dt_alloc_seq();
/* open and read part of an SDfile */
...
/* convert data */
while (NULL_OB != (oseq = dt_convert(conv, iseq))) {
/* loop over molecules */
while (NUL_OB != (obj = dt_next(oseq)))
/* do something with the molecules */
/* destroy output */
dt_dealloc(oseq);
/* read more from SDfile */
...
}
return 0;
}
Sample Conversions - MDl to Daylight
Atom List
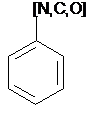
SMARTS: [#7,#6,#8]c1ccccc1
|
Any Atom
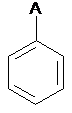
SMARTS: [#0]c1ccccc1
|
Not Aliphatic
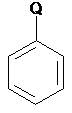
SMARTS: [!#6]c1ccccc1
|
Wild Card Atom
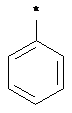
SMILES: *c1ccccc1
|
Lone Pair

SMILES: c1ccncc1
|
R-groups
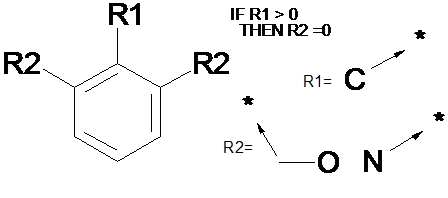
SMARTS: [CO,N]c1cccc([CO,N])c1[C]
|
Isotope
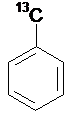
SMILES: [13CH3]c1ccccc1
|
Charge
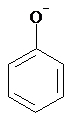
SMILES: [O-]c1ccccc1
|
Implicit Hydrogens
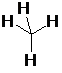
SMARTS: [C&!h0&!h1&!h2&!h3]
|
Stereo Care (none)
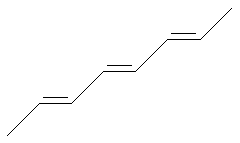
SMILES: C/C=C/C=C/C=C/C
|
Stereo Care (middle)
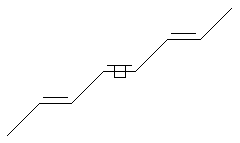
SMILES: CC=C/C=C/C=CC
|
Stereo Care (ends)
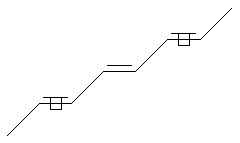
SMILES: C/C=C/C=C\C=C\C
note: implied middle
|
Valence (connections)
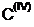
SMARTS: [C&X4]
|
Aromatic Bond
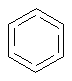
SMARTS: [#6]:1:[#6]:[#6]:[#6]:[#6]:[#6]1
|
Single or Double Bond
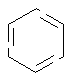
SMARTS: [#6]-,=1[#6]-,=[#6][#6]-,=[#6][#6]1
|
Double or Aromatic Bond

SMARTS: [#6]=,:1-,:[#6]=,:[#6]-,:[#6]=,:[#6]-,:[#6]1
|
Wild Card Bond

SMARTS: [#6]~1~[#6]~[#6]~[#6]~[#6]~[#6]1
|
Chirality

SMILES: C[C@H](N)C(=O)O
|
Chirality (racemic)

SMARTS: C[C&@,@@](N)C(=O)O
|
Stereo (racemic, care all)
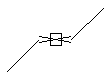
SMARTS: C/,\C=C/C
|
Stereo (racemic ends, care all)
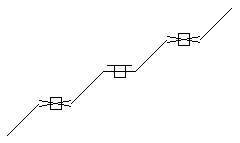
SMARTS: C/,\C=C/C=C/C=C/,\C
|
Stereo (racemic ends, care ends)
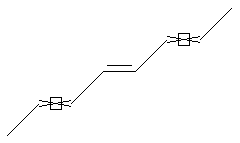
SMARTS: C/,\C=C/C=C/,\C=C/C
|
Stereo (racemic middle, care all)
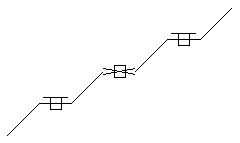
SMARTS: C/C=C/C=C\C=C\C
|
Stereo (racemic middle, care middle and one end)
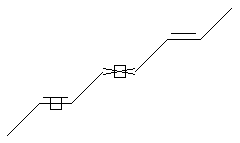
SMARTS: CC=C/,\C=C/C=C/C
|
Stereo (racemic middle, care middle and both one end)
warning: unsupported features
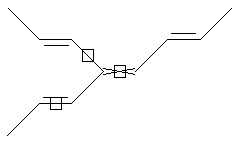
SMARTS: CC=C/,\C=C(/C=C/C)/C=C/C
warning: inconsistent bond directions
SMARTS: CC=C/,\C=C(/C=C/C)\C=C/C
warning: too much symmetry for directional bonds
SMARTS: CC=C/,\C=C(/C)\C=C/C
note: okay
Chain Bond
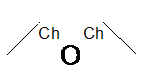
SMARTS: CC-&!@O-&!@CC
| Ring Bond
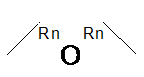
SMARTS: CC-&@O-&@CC
| |
Summary
The Conversion Tools project is designed to meet the needs of chemists wishing to convert chemical information to and from Daylight languages. The product of this project will be a Conversion Toolkit, applications, and an enhanced DayCart system. The initial release will be part of Daylight Software v4.9.
 Daylight Chemical Information Systems, Inc.
Daylight Chemical Information Systems, Inc.
info@daylight.com