Grouping Of Coefficients For The Calculation Of
Inter-Molecular Similarity And Dissimilarity
Using 2D Fragment Bit-strings
J. D. Holliday, C-Y. Hu and P. Willett
Krebs Institute for Biomolecular Research and Department of Information Studies, University of Sheffield, Western Bank, Sheffield S10 2TN, UK
ABSTRACT
This paper compares 22 different similarity coefficients when they are used for searching databases of 2D fragment bit-strings. Experiments with the NCI AIDS and ID Alert databases show that the coefficients fall into several well-marked clusters, in which the members of a cluster will produce comparable rankings of a set of molecules. These clusters provide a basis for selecting combinations of coefficients for use in data fusion experiments. The results of these experiments provide a simple way of increasing the effectiveness of fragment-based similarity searching systems.
INTRODUCTION
Methods for calculating the similarities (or dissimilarities) between pairs, or larger groups, of molecules play an important role in many aspects of chemoinformatics, such as similarity searching [1], property prediction [2], synthesis design [3], virtual screening [4] and molecular diversity analysis [5], inter alia. For example, measures of dissimilarity lie at the heart of many of the algorithms that are used to select libraries of compounds for synthesis and testing that are as structurally diverse as possible. Again, similarity searching involves calculating a measure of structural similarity between a target structure and each of the structures in a database; if the target structure is bioactive, e.g., a weak lead in a drug discovery programme, then inspection of the nearest neighbours resulting from the similarity search can suggest new molecules for biological testing.
The effectiveness of such procedures will be affected by the measure that is used to quantify the degree of similarity or dissimilarity between pairs of structures. There are two principal components to a similarity measure: the representation that is used to characterise the molecules that are to be compared, this often being a set of descriptors such as 2D fragment substructures or sets of calculated physicochemical properties [6,7]; and the similarity coefficient that is used to quantify the degree of resemblance between two such representations. The representation may need some form of pre-processing before the similarity calculation (e.g., descriptor values may need to be weighted or standardised prior to the similarity calculations), thus introducing a third component in such cases. In this paper, we focus on similarity coefficients, in particular their use for measuring similarities between pairs of 2D fragment bit-strings (or fingerprints). These representations provide a very simple encoding of molecular structure but have been found to yield a surprisingly high level of performance in a range of similarity and diversity studies [2, 6-10].
Many different types of similarity coefficient have been described in the literature but most of them can be grouped into three broad classes: distance coefficients, association coefficients and correlation coefficients. Distance coefficients quantify the degree of difference between two objects and have been extensively used in many applications of multivariate statistics (especially where integer-valued or real-valued variables are employed), probably due to the simple geometric interpretation that is attached to many of them (e.g., the Euclidean distance). With a distance coefficient, the greater the degree of similarity between two objects the smaller the value of the coefficient (and vice versa). Association coefficients, conversely, are most commonly used with binary data (i.e., variables denoting the presence or absence of descriptors in an object) and are often normalised to lie within the range of zero (no similarity at all) and unity (identical sets of descriptors). That said, association coefficients can be used with non-binary data, in which case other ranges of values may apply (e.g., the lowerbound of the well-known Tanimoto coefficient is –1/3 when used with such data [1]). Finally, correlation coefficients measure the degree of correlation between the sets of values characterising each of a pair of objects (rather than their more conventional use in multivariate analyses to probe the relationships between pairs of variables). There have been several studies comparing the merits of different chemical similarity measures (see, e.g., [11-13]). It has been found that the Tanimoto coefficient provides a generally effective approach to molecular property prediction and similarity searching, and this coefficient is now widely used for measuring the similarity between pairs of 2D bit-strings (despite some limitations that have recently become apparent [14-16]).
In this paper, we report a comparison of some 22 different similarity coefficients (16 association coefficients, 5 correlation coefficients and 1 distance coefficient) when used with 2D fragment bit-string data, with the aim of identifying some subset of these that exemplify the full range of types of coefficient. Specifically, we have carried out a series of similarity searches using each of the coefficients in turn, clustered them on the basis of the search outputs, and then evaluated the search effectiveness of various combinations of the individual coefficients.
EXPERIMENTAL DETAILS
Assume that the bit-strings XP and XQ denote two molecules P and Q, respectively, and that each of these strings contains a total of n bit positions. Assume further that b and c of these n bits are set to one only in XP and only in XQ, respectively, that a of these n bits are set to one in both XP and XQ, and that d of these n bits are not set in either XP and XQ (so that n = a + b + c + d). Then the various coefficients used here are as shown in Fig. (1), with the first 16 entries being association coefficients, the next five entries being correlation coefficients and the last being a distance coefficient. These coefficients are drawn from the extensive review by Ellis et al. [17], who also include several other distance coefficients. However, all but one of these (the Bray-Curtis distance) are completely monotonic, i.e., will result in identical rankings of a set of molecules in response to a query, and we have hence used just one of them, the mean Manhattan Distance, in our experiments1. The remaining distance coefficient, the Bray-Curtis, has also been omitted as it proved to be the complement of the Dice Coefficient (coefficient-2 in Fig. (1)). The reader is referred to the review by Ellis et al. for a detailed discussion of the origins of the various coefficients that we have tested here [17].
The principal dataset used in our experiments was the 1999 version of the National Cancer Institute’s AIDS database [18], which contains compounds that have been tested for anti-HIV activity. Salts and duplicate structures were removed from this file to give a total of 37,124 compounds, 294 of which have been confirmed as showing strong activity. These molecules were represented by their UNITY 2D fragment bit-strings [19], hashed fingerprint representations which encode the 2D structural features present in the molecule. These features include fragment-based sequences of length two to six, five- and six-membered ring systems, and counts of non-carbon atoms. The bit-strings for all of the compounds were used in all of the experiments discussed below. 60 of the strongly active molecules were chosen from the file to act as the target structures for database searching.
CLUSTERING OF RANKINGS
A similarity search of the AIDS database was carried out using each of the 22 similarity measures shown in Fig. (1). The results of each search were ranked in decreasing order of the calculated similarity coefficient (or the complement in the case of the distance coefficient). The rankings for two searches can be compared by counting the number of compounds in common in the top t structures, (where t has been chosen as 50, 100, 200 and 400 for this experiment); then the dissimilarity value Dij between searches using coefficients i and j is defined to be
![]() ,
,
where cij is the number of common structures in the top t ranking positions. It is hence possible to generate a t´ t dissimilarity matrix for each target structure, illustrating the relationships between pairs of similarity coefficients for that target at the chosen value of t. Three different agglomerative hierarchical clustering methods (single linkage, complete linkage and group-average) were then applied to each such dissimilarity matrix; the clustering of similarity coefficients to identify inter-coefficient relationships was first suggested by Hubaleck in the context of fungal species extracted from birds and bird nests [20]. The dendrograms obtained here, using the first target structure and with t=50, are shown in Figs. (2-4).
After clustering, Mojena’s stopping rule [21] was used to determine a stopping level at which to partition each of the resulting dendrograms into appropriate groups. Here, a hierarchy on m objects is partitioned at that level for which
![]()
where a 0, a 1….a m-1 are the dissimilarity levels at which each successive agglomeration takes place, these corresponding to stages with m, m-1….1 clusters. The terms m and s are the mean and unbiased standard deviation, respectively, of the a values, and k is a constant (which we set to 1.25 as suggested by Milligan and Cooper [22]).
By using Mojena’s stopping rule, the dendrograms of Figs. (2-4) are all partitioned to give (the same) three groups. These are (the numbers in the groups correspond to the coefficients in Fig. (1)):
Group A : {1 2 4 5 11 12 15 17 20 21}
Group B : {3 13 16 18 19}
Group C : {6 7 8 9 10 14 22}
The same procedure was carried out for all classifications (60 different target structures and three different clustering methods for each target) and we then combined the resulting 180 sets of groupings to identify groups of coefficients that are always clustered together. This overall grouping contained a total of 11 groups as follows:
Group A : {3}
Group B : {11}
Group C : {14}
Group D : {16}
Group E : {18}
Group F : {21}
Group G : {1 2 4 5}
Group H : {6 7 8 9 10 22}
Group I : {12 15}
Group J : {13 19}
Group K : {17 20}
Thus, for example, coefficients 1, 2, 4 and 5 clustered together in all 180 tests, as did 6, 7, 8, 9, 10 and 22, etc. The reader should note that the presence of several coefficients in a group does not imply that all of the coefficients are strictly monotonic; however, it does mean that the sets of nearest neighbour compounds resulting from their use are very similar, and it is these sets, rather than the precise rankings, that are normally required in database searching applications
Having identified the groups of closely related coefficients, we can then assess the relationships between these groups. These are illustrated in Table 1, which contains the number of times that each of the 11 groups above clustered with one of the other 10 groups over the whole set of 180 classifications. It will be seen that there are some very strong relationships between the groups, e.g., Groups G and I (containing coefficients {1,2,4,5} and {12,15}, respectively) clustered together in no less than 178 of the 180 tests. If we were to use 170 co-occurrences, then these two groups would be joined by Groups B, F and K to give the grouping {1,2,4,5,11,12,15,17,20,21}. Thus far, we have considered only the groupings obtained with t=50, but very similar results are obtained if t is set to 100, 200 or 400, rather than 50 as in Table 1. In fact, although there are 11 groups using the criteria discussed above, the Mojena stopping rule normally identified just three or four groups of coefficients in the great majority of the experiments that were carried out, whatever the target structure, clustering method or value of t: one consisting mainly of coefficients from the set {1,2,4,5,11,12,15,17,20,21}; one consisting mainly of coefficients from the set {6,7,8,9,10,22}; and one consisting of the others, although coefficient 3 was often found on its own.
To ensure that these results were not determined by the specific characteristics of the AIDS database and of the target structures that were used, a comparable series of experiments was carried out using 11607 structures for which biological activities have been reported in the literature and which were entered into the IDAlert database during 1992-96 [23]. 60 molecules were again selected at random from this file and used as target structures for similarity searching. The search rankings were analysed as described previously and yielded groupings very similar to those obtained with the AIDS data. For example, using a threshold of 170 with t=50, the largest group contained coefficients {1,2,4,5,12,15,17,20}, i.e., identical to that obtained with the AIDS dataset with the exception of coefficients 11 and 21, both which here were in clusters on their own. This similarity of behaviour was observed for searches using all four values of t (50, 100, 200 and 400).
It hence seems reasonable to conclude that there are groups of coefficients that tend to produce analogous rankings when used for similarity searching in chemical databases. This conclusion is hardly surprising when one considers the actual formulae in Fig. (1). For example, we have noted that coefficients 6-10 occur together, and they all involve the term a+d in the numerator of their defining expression; coefficient 22 (mean Hamming) also co-occurs and subsequent inspection of its formula revealed that it is actually the complement of coefficient 6 (Simple Match), as the term b+c in the Hamming numerator 22 is simply n-(a+d). At the same time, there are coefficients that are markedly different from each other, with the sets of top-ranked nearest neighbours having less than 5% of the molecules in common. These differences are demonstrated in Table 1; we have already noted that coefficient 3 is a frequent outlier, but coefficients 14, 16 and 18 also have notably fewer co-occurrences (except with each other) than the other coefficients considered here.
DATA FUSION
An inspection of the literature on chemical similarity and dissimilarity reveals that while there are many different similarity measures, most published studies have considered the use of only a single measure. Even where this is not the case, multiple measures have typically been employed only as the input to a comparative study that seeks to identify the "best" measure, using some quantitative performance criterion. Such comparisons are limited in that they assume, normally implicitly, that there is some specific type of structural feature, weighting scheme or whatever that is uniquely well suited to describing the type(s) of biological activity that are being sought for in a similarity search. The assumption cannot be expected to be generally valid, given the multi-faceted nature of biological activities, and there has thus recently been interest in the use of data fusion or consensus scoring methods [24-27]. For example, Charifson et al. have discussed combining rankings based on different scoring functions for docking searches [26], an idea that is embodied in the CSCORE software package [19]. Such combined rankings generally give better results than use of an individual scoring function or similarity measure, and small-scale experiments suggest that the more rankings that are combined, the better the final results [27]. However, if many different similarity measures are available, it may be too time-consuming to use all of them, in which case it seems appropriate to ensure that as diverse a range of types of measure are employed (in much the same way as one focuses on structural diversity when selecting database subsets for biological testing). Here, we use the results of our clustering experiments to provide a rational basis for selecting similarity coefficients for data fusion.
We took as our starting point the groups of coefficients identified previously, and then chose one coefficient from each group as representative of the different types of similarity measure: in many of the groups, of course, there was only a single coefficient to choose. The coefficients chosen from groups A-K were as follows: 3 (Russell/Rao), 11 (Baroni-Urbani/Buser), 14 (Forbes), 16 (Simpson), 18 (Yule), 21 (Dennis), 1 (Jaccard/Tanimoto), 6 (Simple Match), 12 (Ochiai/Cosine), 13 (Kulczynski(2)) and 20 (Stiles), respectively. A set of 20 active molecules was then randomly chosen from the AIDS database and each of these was used as the target structure for a similarity search of the database, using each of the chosen coefficients in turn. These target structures are shown in Figure (5) to illustrate the range of structural types considered in the searches.
We have already noted that most of the classifications suggested the presence of three groups of coefficients, and fusion was hence carried out for each target structure by selecting the rankings for three coefficients at a time. These rankings were merged by summing the rank-positions for each compound to give a score that formed the basis for a new, final ranking (this is the SUM fusion rule that was found to be the most effective in the study of Ginn et al. [27]). Each ranking, whether from one of the 11 original individual coefficients or from one of the 165 (3C11) combinations of coefficients, was then inspected to identify the number of active molecules in the top 400 positions; this number was taken as the effectiveness of that search, and hence of the effectiveness of the combination of coefficients, or the individual coefficient, that was used in the search.
For each target structure, the combinations were sorted into decreasing order of the number of actives retrieved and assigned an ordinal value from 1 (best ranking) down to 165 (worst ranking); combinations with equal performance were assigned the mean ordinal value (i.e., if the third, fourth and fifth best combinations all retrieved the same number of actives then they are all assigned 4). The overall performance of each combination was then determined by the sum of ordinal values across all 20 target structures, with the results in Table 2. The upper part of this table contains the 10 best combinations, and the bottom part the 10 worst combinations. If all of the combinations performed at the same level, then the summed value would be 1660, i.e., the median of 1,2,3….164, 165, multiplied by 20 (the number of searches); the results in the table hence demonstrate the substantial differences in search effectiveness that can arise as the result of a good, or a bad, choice of coefficients for carrying out data fusion. The table also contains the median numbers of actives retrieved when averaged over the 20 searches, this again demonstrating clearly the substantial performance differences that exist.
The top row of Table 2 represents the best single combination (on the basis of our chosen measure of search effectiveness) and consists of the following three coefficients: the Russell/Rao coefficient; the Simple Match coefficient; and the Stiles coefficient. Other coefficients figuring prominently are Jaccard/Tanimoto and Kulczynski(2). It is worth mentioning the fifth-ranked combination (Jaccard/Tanimoto, Russell/Rao and Simple Match) as Dixon and Koehler have recently recommended the combination of Tanimoto and Hamming (which we have seen is the complement of the Simple Match) for similarity searching [16]. The worst single combination involves the Forbes coefficient, the Dennis coefficient and the Simple Match coefficient, and all three of these figure prominently in the bottom part of Table 2; Dennis indeed occurs in every one of them. It is surprising to see Simple Match appearing so frequently here, given that it also figured prominently at the top of the ranking. In fact, an analysis of all of the combinations involving this coefficient shows that it appears in combinations that are spread throughout the ranked list of 165 combinations for each query, as we now demonstrate. Each individual coefficient, c, will occur in 45 different combinations with pairs of other coefficients; then the sum of the positions in the ranked list for each of these 45 combinations will give an overall figure-of-merit for coefficient c. These figures are shown in Table 3, from which we see that the best individual coefficients (as manifested by their behaviour in the data fusion experiments) are Russell/Rao, Jaccard/Tanimoto and then Ochai/Cosine, while the worst are Dennis (which occurred in all of the 45 lowest ranked combinations), Forbes and then Simpson. The frequent high and low appearances in Table 2 of Simple Match means that it occurs around the middle of the ranking in Table 3.
Previous studies of data fusion have suggested that appropriately combined rankings can be consistently superior to individual rankings, and this is certainly the case here. Table 4 lists numbers of actives retrieved in each of the 20 searches for the 11 individual coefficients and for the best three-way combination (i.e., Russell/Rao, Simple Match and Stiles). Each element of the main body of the table also contains an enrichment factor. Assume that an individual coefficient retrieves Ni actives, and the best three-way combination Nc actives, in the top 400 rank positions. Then the enrichment is defined to be
![]()
The median values for numbers of actives retrieved and for the enrichment are listed at the bottom of the table, where it will be seen that our best 3-coefficient combination is superior, when averaged over the 20 searches, to all of the individual coefficients (in that the median number of actives is greater than the numbers for the individual rankings, and in that all of the enrichment factors are positive).
The focus of this section of the paper is the performance of the fused similarity coefficients, but the median values in Table 4 enable one to compare the individual coefficients. If one takes the numbers of actives retrieved, then the order is
1>11=12>13>20=21>18>6>3>16>14,
while for the enrichments the order is
13>1=12>3>11>20>21>6>18>16>14.
There is a fair degree of commonality with the ranking of the fused combinations discussed above, albeit with some obvious discrepancies - specifically coefficient 3 (Russell/Rao) in the actives retrieved and 21 (Dennis) in both actives retrieved and enrichment - when compared with the combination rankings in Table 3. In general, then, if an individual coefficient performs well, then it is likely (but not guaranteed) to perform well when combined with other effective coefficients; in a similar vein, poorly performing individual coefficients are not expected to do well when combined with other poorly performing coefficients.
The clustering experiments suggested the existence of three groups of coefficients, and hence the use of 3-way fusions in the experiments thus far. To confirm the optimality of this choice, a further study was carried out in which all possible combinations of any number of coefficients were investigated. The same method of fusion, the merging of rank-positions, was used as for the 3C11 combinations, but this time we chose all nC11 combinations for values of n from 2 to 11. The number of combinations for each value of n is shown in the second column of Table 5. We used the same twenty actives as targets and merged the respective rankings. Performance was measured in two ways: firstly, using the sum of ordinal performance values as in the previous study; and, secondly, using the number of actives in the top 400 rank positions summed across all 20 searches. The best combinations based on these two performance measures for all values of n are shown in Table 5. This table also includes values for the performance of the single coefficients (i.e., n=1) and, for each value of n, indicates the sum of actives retrieved by the best combination and the mean and median values for the sum of actives across all combinations. It will be seen that the best combination is either 3 (Russell/Rao) and 6 (SimpleMatch) or 3, 6 and 20 (Stiles), i.e., the best 3-way combination used previously. Both of these combinations retrieve the same number of actives (843) and are a considerable improvement on the performance of the best single coefficient (803 actives).
The median number of actives retrieved appears to fall considerably for combinations of greater than six coefficients, reflecting the increasing influence of the poorly-performing coefficients such as 21 (Dennis). The value for the sum of actives for the best combination remains high for nearly all values of n, while the mean decreases steadily. When compared to the median, this appears to indicate that there is a set of combinations which perform comparably well, and most combinations taken from this set (which appears to include Russell/Rao, SimpleMatch, Stiles, Jaccard/Tanimoto, Ochiai/Cosine, Baroni-Urbani/Buser, and Kulcynski(2)), should give an acceptable level of performance..
CONCLUSIONS
In this paper we have compared a range of similarity coefficients when they are used for similarity and dissimilarity searching in databases of chemical compounds represented by 2D fragment bit-strings. Our experiments with the NCI AIDS and IDAlert databases demonstrate clearly that the coefficients fall into several well-marked clusters, in which the members of a cluster will produce comparable sets of nearest neighbours in similarity searches for a given target compound. We have then used these clusters as the basis for a data fusion study in which we have combined the results of similarity searches based on three different coefficients to produce new, fused rankings. These fused rankings are generally more effective, in terms of retrieving bioactive molecules, than rankings based on just a single coefficient, if an appropriate combination of coefficients is chosen for the fusion.
This finding provides a simple way of enhancing the performance of existing systems for fragment-based similarity searching. Current systems compare the bit-string for a target structure with the bit-strings of each of the molecules in a database, using the common and non-common bits in each such comparison to calculate a value for (typically) the Tanimoto coefficient. Our results suggest that if these common bits are additionally used to calculate the values of other coefficients, then the resulting ranking will contain a larger number of high-ranked active compounds than if just the Tanimoto is used. These additional coefficient values can be calculated at negligible computational cost (since the bit-string comparisons have already been performed for the Tanimoto calculation): the use of data fusion as advocated here hence results in an increase in search effectiveness with only a very slight decrease in search efficiency.
Clearly this work could be considerably extended, for example by using other types of 2D fragment bit-string, by varying the number of coefficients involved in each combination ranking, and by using datasets for which quantitative, rather than qualitative, bioactivity data are available. Summarising the results we have obtained thus far, our experiments certainly support the continued popularity of the Jaccard/Tanimoto coefficient for chemoinformatic applications; in addition, researchers might also usefully consider, either alone or in combination, the Russell/Rao, Baroni-Urbani/Buser, SimpleMatch, Stiles, Ochiai/Cosine, and Kulczynski(2) coefficients.
The final point we wish to make is that our findings are specific to 2D fragment bit-strings; and are not necessarily applicable to other types of representation. For example, the HQSAR system [19] uses a representation that involves frequencies of occurrence of individual fragments, rather than just their presence or absence. Other descriptors result in real-valued representations, such as the sets of topological indices produced by MOLCONN-Z [28] or the field values produced by 3D descriptors such as COMFA [19]. Such molecular descriptors may require the use of standardisation and are also hospitable to a larger range of similarity coefficients than used here (where many of the coefficients that one might consider are rendered completely monotonic by the use of binary representations). The use of data fusion with such descriptors provides another obvious area for future experimentation.
ACKNOWLEDGEMENTS
We thank the KC Wong Education Foundation for funding, Tripos Inc. for software support and Current Drugs Limited for provision of the IDAlert database. The Krebs Institute for Biomolecular Research is a designated biomolecular sciences centre of the Biotechnology and Biological Sciences Research Council.
REFERENCES
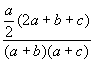
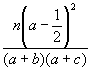
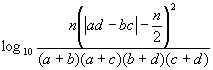
Figure 1. Association coefficients (1-16), correlation coefficients (17-21) and distance coefficient (22) used in the experiments.
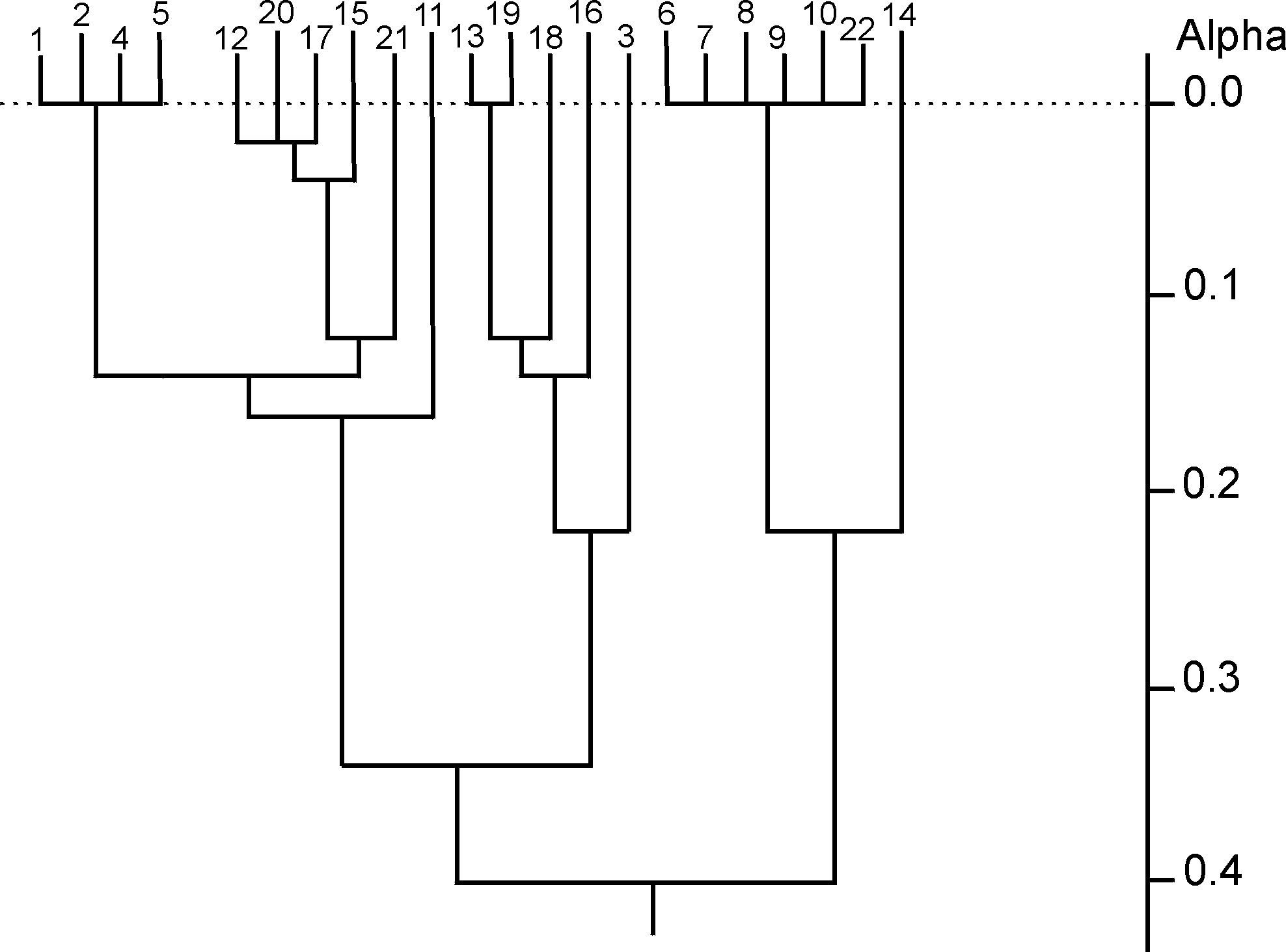
Figure 2. Dendrogram for single linkage clustering of first target structure dissimilarity matrix;
a j+1= 0.274 implies stopping at level 19 by Mojena's rule(k=1.25)
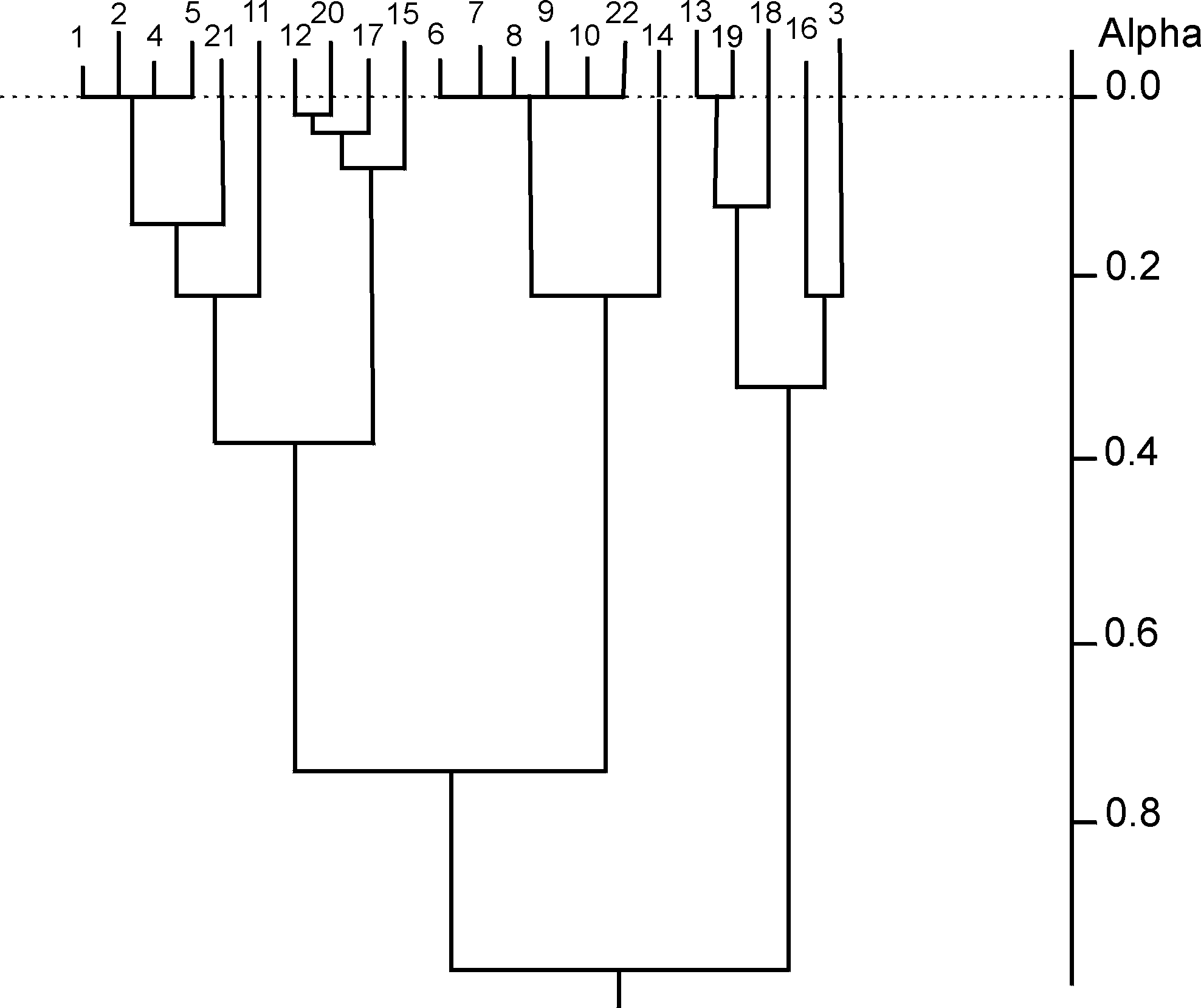
Figure 3. Dendrogram for complete linkage clustering of first target structure dissimilarity matrix; a j+1= 0.581 implies stopping at level 19 by Mojena's rule(k=1.25)
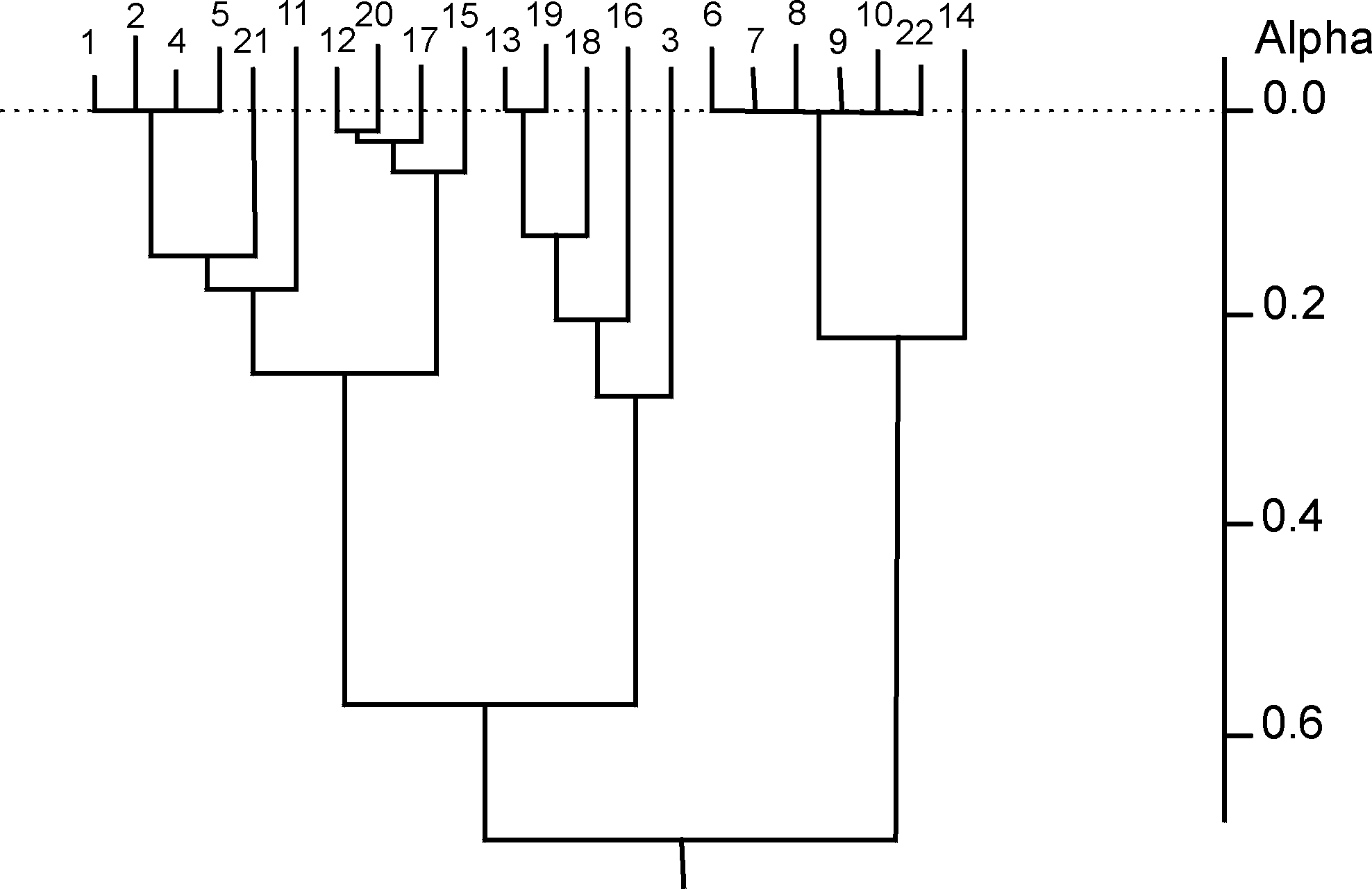
Figure 4. Dendrogram for group average linkage clustering of first target structure dissimilarity matrix; a j+1= 0.366 implies stopping at level 19 by Mojena's rule(k=1.25)
 Figure
5. The 20 actives from Aids database used as target structures
used for data fusion.
Figure
5. The 20 actives from Aids database used as target structures
used for data fusion.
|
A 3 |
B 11 |
C 14 |
D 16 |
E 18 |
F 21 |
G {1,2,4,5} |
H {6,7,8,9,10,22} |
I {12,25} |
J {13,19} |
K {17,20} |
|
|
A |
0 |
||||||||||
|
B |
10 |
0 |
|||||||||
|
C |
0 |
19 |
0 |
||||||||
|
D |
52 |
4 |
112 |
0 |
|||||||
|
E |
18 |
27 |
123 |
126 |
0 |
||||||
|
F |
10 |
174 |
15 |
4 |
27 |
0 |
|||||
|
G |
13 |
172 |
15 |
4 |
27 |
174 |
0 |
||||
|
H |
9 |
142 |
28 |
5 |
32 |
141 |
138 |
0 |
|||
|
I |
14 |
170 |
15 |
5 |
28 |
172 |
178 |
137 |
0 |
||
|
J |
14 |
162 |
16 |
11 |
37 |
164 |
167 |
136 |
169 |
0 |
|
|
K |
10 |
175 |
15 |
4 |
27 |
177 |
177 |
138 |
175 |
167 |
0 |
Table 1: Relationships between pairs of groups of coefficients. Each element in the table lists the number of times that one particular group of coefficients occurred with another particular combination (see text).
|
Combination |
Ordinal Value |
Median Actives |
|
3,6,20 |
993.5 |
40.5 |
|
3,12,13 |
1032.5 |
37.5 |
|
1,3,13 |
1047.0 |
37.5 |
|
1,3,12 |
1054.0 |
37.5 |
|
1,3,6 |
1065.5 |
40.0 |
|
3,6,12 |
1082.0 |
40.0 |
|
3,6,11 |
1089.0 |
40.0 |
|
3,11,18 |
1093.0 |
37.0 |
|
3,6,13 |
1105.5 |
40.0 |
|
1,13,18 |
1108.0 |
37.5 |
|
11,18,21 |
3748.0 |
2.0 |
|
13,14,21 |
3756.0 |
1.0 |
|
6,11,21 |
3764.0 |
1.0 |
|
14,16,21 |
3854.0 |
1.0 |
|
6,20,21 |
3856.0 |
1.0 |
|
6,18,21 |
3881.5 |
1.0 |
|
6,16,21 |
3956.0 |
1.0 |
|
14,18,21 |
3964.5 |
1.0 |
|
14,20,21 |
3964.5 |
1.0 |
|
6,14,21 |
3977.5 |
1.0 |
Table 2: Best (top 10 rows) and worst (bottom 10 rows) combinations of coefficients for similarity searching in the AIDS database. The second column gives the sum of rank positions over the set of 20 target structures, and the third column the median number of actives retrieved when averaged over these 20 searches.
|
Coefficient |
Sum Of Rank Positions |
|
3 |
1964 |
|
1 |
2981 |
|
12 |
3191 |
|
13 |
3314 |
|
11 |
3468 |
|
20 |
3548 |
|
18 |
3733 |
|
6 |
3853 |
|
16 |
3912 |
|
14 |
4686 |
|
21 |
6435 |
Table 3. Sum of rank positions in Table 2 for combinations involving each of the 11 selected individual coefficients.
|
Best Fusion |
Coefficient |
|||||||||||
|
1 |
3 |
6 |
11 |
12 |
13 |
14 |
16 |
18 |
20 |
21 |
||
|
34 |
33 3.0 |
34 0.0 |
34 0.0 |
33 3.0 |
33 3.0 |
33 3.0 |
28 21.4 |
34 0.0 |
34 0.0 |
33 3.0 |
33 3.0 |
|
|
41 |
42 -2.4 |
32 28.1 |
42 -2.4 |
42 -2.4 |
42 -2.4 |
42 -2.4 |
31 32.3 |
31 32.3 |
38 7.9 |
42 -2.4 |
42 -2.4 |
|
|
1 |
1 0.0 |
2 -50.0 |
1 0.0 |
1 0.0 |
1 0.0 |
1 0.0 |
1 0.0 |
1 0.0 |
1 0.0 |
1 0.0 |
1 0.0 |
|
|
8 |
7 14.3 |
12 -33.3 |
2 300.0 |
4 100.0 |
8 0.0 |
9 -11.1 |
2 300.0 |
12 -33.3 |
6 33.3 |
4 100.0 |
4 100.0 |
|
|
5 |
5 0.0 |
5 0.0 |
5 0.0 |
5 0.0 |
5 0.0 |
5 0.0 |
4 25.0 |
4 25.0 |
5 0.0 |
5 0.0 |
5 0.0 |
|
|
79 |
73 8.2 |
70 12.9 |
63 25.4 |
69 14.5 |
72 9.7 |
70 12.9 |
34 132.4 |
61 29.5 |
65 21.5 |
68 16.2 |
66 19.7 |
|
|
3 |
3 0.0 |
4 -25.0 |
3 0.0 |
3 0.0 |
3 0.0 |
3 0.0 |
3 0.0 |
3 0.0 |
3 0.0 |
3 0.0 |
3 0.0 |
|
|
73 |
68 7.4 |
70 4.3 |
61 19.7 |
66 10.6 |
68 7.43 |
70 4.3 |
20 265.0 |
35 108.6 |
58 25.9 |
63 15.9 |
62 17.7 |
|
|
40 |
36 11.1 |
35 14.3 |
34 17.7 |
36 11.1 |
35 14.3 |
34 17.7 |
20 100.0 |
28 42.9 |
35 14.3 |
34 17.7 |
34 17.7 |
|
|
42 |
40 5.0 |
33 27.3 |
34 23.5 |
39 7.7 |
40 5.0 |
39 7.7 |
21 100.0 |
29 44.8 |
36 16.7 |
38 10.5 |
38 10.5 |
|
|
78 |
75 4.0 |
69 13.0 |
69 13.0 |
74 5.4 |
75 4.0 |
75 4.0 |
39 100.0 |
50 56.0 |
65 20.0 |
71 9.9 |
69 13.0 |
|
|
14 |
14 0.0 |
15 -6.7 |
16 -12.5 |
15 -6.7 |
15 -6.7 |
16 -12.5 |
16 -12.5 |
17 -17.7 |
15 -6.7 |
15 -6.7 |
15 -6.7 |
|
|
78 |
75 4.0 |
67 16.4 |
71 9.9 |
74 5.4 |
73 6.9 |
72 8.3 |
41 90.2 |
50 56.0 |
66 18.2 |
72 8.3 |
71 9.9 |
|
|
5 |
2 150.0 |
17 -70.6 |
2 150.0 |
2 150.0 |
3 66.7 |
5 0.0 |
2 150.0 |
16 -68.8 |
3 66.7 |
2 150.0 |
2 150.0 |
|
|
18 |
17 5.9 |
22 -18.2 |
12 50.0 |
15 20.0 |
17 5.9 |
16 12.5 |
11 63.6 |
11 63.6 |
12 50.0 |
13 38.5 |
13 38.5 |
|
|
74 |
68 8.8 |
61 21.3 |
66 12.1 |
68 8.8 |
67 10.5 |
67 10.5 |
28 164.3 |
33 124.2 |
64 15.6 |
66 12.1 |
66 12.1 |
|
|
15 |
16 -6.3 |
14 7.1 |
16 -6.3 |
17 -11.8 |
16 -6.3 |
16 -6.3 |
16 -6.3 |
15 0.0 |
16 -6.3 |
17 -11.8 |
17 -11.8 |
|
|
78 |
74 5.4 |
70 11.4 |
71 9.9 |
73 6.8 |
74 5.4 |
74 5.4 |
41 90.2 |
50 56.0 |
65 20.0 |
71 9.9 |
71 9.9 |
|
|
83 |
73 13.7 |
69 20.3 |
71 16.9 |
72 15.3 |
72 15.3 |
72 15.3 |
25 232.0 |
33 151.5 |
69 20.3 |
72 15.3 |
71 16.9 |
|
|
74 |
71 4.2 |
69 7.2 |
65 13.9 |
68 8.8 |
71 4.2 |
70 5.7 |
34 117.7 |
54 37.0 |
56 32.1 |
65 13.9 |
65 13.6 |
|
|
Median values |
||||||||||||
|
40.5 |
38 |
33.5 |
34 |
37.5 |
37.5 |
36.5 |
20.5 |
30 |
35.5 |
36 |
36 |
|
|
4.6 |
7.2 |
12.6 |
7.3 |
4.6 |
4.1 |
95.1 |
34.7 |
17.4 |
10.2 |
11.3 |
||
Table 4. Relative performance of the best 3-coefficient fusion combination (Russell/Rao, SimpleMatch and Stiles, in the left-hand column) compared with the use of individual coefficients. The results for the 20 target structures are shown in terms of the number of actives retrieved and the percentage enrichment (in italics), based on the top 400 positions in each search ranking. The median retrieval and enrichment over all 20 targets is shown in the bottom row.
|
n |
nC11 |
Best combination based on sum of ordinal values |
Best combination based on sum of actives retrieved |
Sum of actives retrieved |
Mean and median sum of actives retrieved |
|
1 |
11 |
- |
12 or 13 |
803 |
714 755 |
|
2 |
55 |
3,6 |
3,6 |
843 |
612.2 754 |
|
3 |
165 |
3,6,20 |
3,6,20 |
843 |
563.6 749 |
|
4 |
330 |
1,3,13,20 |
1,3,12,20 or 1,3,13,20 |
841 |
504.7 724.5 |
|
5 |
462 |
3,13,16,18,20 |
1,3,12,13,20 |
833 |
441.8 721 |
|
6 |
462 |
1,3,12,13,16,18 |
1,3,11,12,13,20 |
824 |
379.8 75.5 |
|
7 |
330 |
1,3,12,13,16,18,20 |
1,3,12,13,16,18,20 |
818 |
320.5 79 |
|
8 |
165 |
1,3,11,12,13,16,18,20 |
1,3,11,12,13,16,18,20 |
814 |
264.8 89 |
|
9 |
55 |
1,3,6,11,12,13,16,18,20 |
1,3,6,11,12,13,16,18,20 |
794 |
221.4 103 |
|
10 |
11 |
All but 21 |
All but 21 |
765 |
170.4 109 |
|
11 |
1 |
All |
All |
157 |
157 157 |
Table 5. Best combinations of fused coefficients based on the sum of ordinal values and on sum of actives. The fourth column gives the sum of actives retrieved by the best combination and the fifth column gives the mean and median values for the sum of actives across all combinations of n coefficients.